Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for AHR_ARNT2
Z-value: 2.04
Transcription factors associated with AHR_ARNT2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
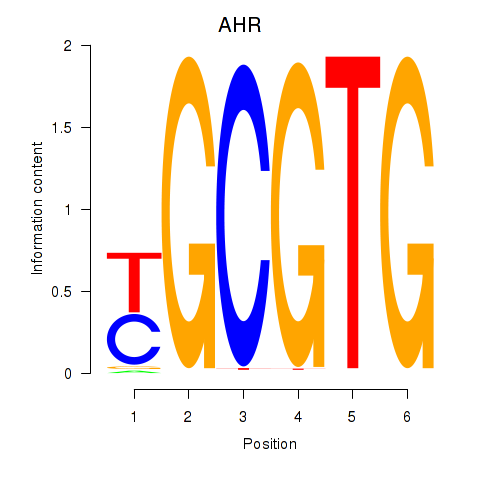
AHR
|
ENSG00000106546.14 | aryl hydrocarbon receptor |
|
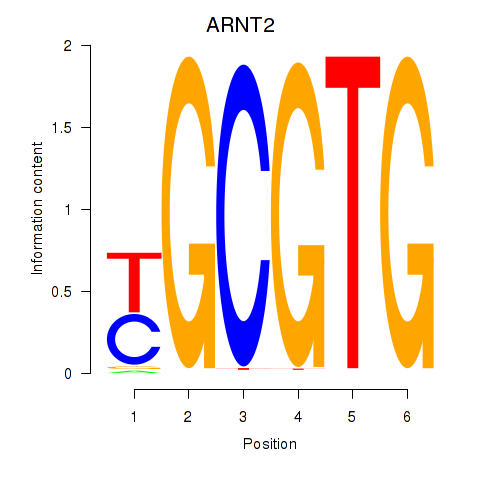
ARNT2
|
ENSG00000172379.21 | aryl hydrocarbon receptor nuclear translocator 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARNT2 | hg38_v1_chr15_+_80441229_80441313 | 0.72 | 4.2e-06 | Click! |
| AHR | hg38_v1_chr7_+_17298642_17298658 | -0.07 | 7.0e-01 | Click! |
Activity profile of AHR_ARNT2 motif
Sorted Z-values of AHR_ARNT2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AHR_ARNT2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.4 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 3.5 | 10.4 | GO:0008355 | olfactory learning(GO:0008355) |
| 1.5 | 5.9 | GO:0061163 | endoplasmic reticulum polarization(GO:0061163) actin filament bundle retrograde transport(GO:0061573) actin filament bundle distribution(GO:0070650) |
| 1.4 | 4.3 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 1.3 | 3.9 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 1.3 | 3.8 | GO:0021586 | pons maturation(GO:0021586) |
| 1.2 | 4.7 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 1.0 | 5.2 | GO:0070982 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 1.0 | 3.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 1.0 | 7.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 1.0 | 2.9 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.9 | 2.8 | GO:2000374 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) regulation of oxygen metabolic process(GO:2000374) |
| 0.9 | 3.5 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.9 | 18.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.8 | 3.9 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.8 | 2.3 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.8 | 2.3 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.8 | 2.3 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.8 | 8.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.7 | 5.2 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.7 | 2.2 | GO:1903452 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.7 | 4.8 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.7 | 10.8 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.7 | 9.3 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.7 | 3.3 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.7 | 2.0 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 0.6 | 1.9 | GO:0070446 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.6 | 3.7 | GO:0042819 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) |
| 0.6 | 1.8 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.6 | 2.4 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.6 | 3.0 | GO:0070352 | positive regulation of white fat cell proliferation(GO:0070352) |
| 0.6 | 23.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.5 | 2.7 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.5 | 2.2 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.5 | 1.5 | GO:0007309 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.5 | 4.6 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.5 | 2.5 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.5 | 5.5 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.5 | 0.5 | GO:1902959 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.5 | 28.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.5 | 6.8 | GO:0003360 | brainstem development(GO:0003360) |
| 0.5 | 1.4 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.5 | 8.9 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.5 | 1.4 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.4 | 1.3 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.4 | 1.3 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.4 | 6.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.4 | 3.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.4 | 1.2 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.4 | 0.4 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.4 | 2.0 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.4 | 5.2 | GO:0015820 | leucine transport(GO:0015820) |
| 0.4 | 2.4 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.4 | 1.2 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.4 | 1.6 | GO:0031106 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.4 | 0.8 | GO:0042704 | uterine wall breakdown(GO:0042704) |
| 0.4 | 3.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.4 | 1.9 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.4 | 2.7 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.4 | 5.7 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.4 | 1.5 | GO:1905075 | positive regulation of relaxation of muscle(GO:1901079) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.4 | 1.1 | GO:2000851 | positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.4 | 1.5 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.4 | 2.2 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.4 | 3.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.4 | 1.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.4 | 9.9 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.4 | 2.5 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.4 | 3.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.3 | 3.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.3 | 0.7 | GO:0071464 | cellular response to hydrostatic pressure(GO:0071464) |
| 0.3 | 1.0 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.3 | 0.7 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.3 | 1.0 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.3 | 4.6 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.3 | 2.9 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.3 | 1.0 | GO:0070662 | mast cell proliferation(GO:0070662) |
| 0.3 | 1.3 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.3 | 1.2 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.3 | 4.0 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.3 | 0.9 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.3 | 1.5 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.3 | 1.2 | GO:1903892 | negative regulation of ATF6-mediated unfolded protein response(GO:1903892) |
| 0.3 | 0.3 | GO:1904782 | negative regulation of glutamate receptor signaling pathway(GO:1900450) negative regulation of NMDA glutamate receptor activity(GO:1904782) |
| 0.3 | 0.9 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.3 | 0.6 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.3 | 1.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.3 | 12.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 3.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.3 | 2.8 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.3 | 1.4 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.3 | 4.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.3 | 1.6 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) |
| 0.3 | 0.8 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.3 | 6.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.3 | 2.7 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.3 | 2.7 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.3 | 1.6 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.3 | 3.4 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.3 | 1.3 | GO:0048690 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.3 | 0.8 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.3 | 13.2 | GO:0007616 | long-term memory(GO:0007616) |
| 0.3 | 4.8 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.3 | 1.5 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.3 | 2.5 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.3 | 0.5 | GO:0032417 | regulation of sodium:proton antiporter activity(GO:0032415) positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.2 | 1.5 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 0.5 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.2 | 2.0 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 0.7 | GO:1903762 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.2 | 0.7 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.2 | 0.7 | GO:0006683 | galactosylceramide catabolic process(GO:0006683) |
| 0.2 | 0.7 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.2 | 2.1 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 | 1.2 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.2 | 1.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.2 | 3.9 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.2 | 0.7 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.2 | 0.9 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.2 | 2.7 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.2 | 1.8 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.2 | 0.9 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.2 | 3.1 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 1.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 0.9 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.2 | 0.6 | GO:0097698 | telomere maintenance via base-excision repair(GO:0097698) |
| 0.2 | 0.9 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.2 | 0.6 | GO:0035724 | CD24 biosynthetic process(GO:0035724) activation of meiosis involved in egg activation(GO:0060466) negative regulation of monocyte extravasation(GO:2000438) regulation of CD24 biosynthetic process(GO:2000559) positive regulation of CD24 biosynthetic process(GO:2000560) |
| 0.2 | 1.3 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.2 | 0.8 | GO:0072560 | type B pancreatic cell maturation(GO:0072560) |
| 0.2 | 0.6 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 1.5 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 2.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 2.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 11.0 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 0.8 | GO:1904980 | positive regulation of endosome organization(GO:1904980) |
| 0.2 | 2.0 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 0.4 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.2 | 0.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 1.0 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.2 | 2.1 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 | 0.2 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.2 | 1.7 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.2 | 1.7 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 1.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.2 | 0.6 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.2 | 8.2 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.2 | 1.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 0.7 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.2 | 0.6 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.2 | 7.9 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.2 | 0.7 | GO:0072021 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.2 | 3.2 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.2 | 1.8 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 | 0.9 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 0.5 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.2 | 2.6 | GO:1903056 | regulation of melanosome organization(GO:1903056) |
| 0.2 | 0.9 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.2 | 0.5 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.2 | 0.5 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.2 | 0.7 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.2 | 1.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.2 | 2.0 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.2 | 1.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 1.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 1.5 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 0.5 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.2 | 3.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 0.5 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.2 | 1.8 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.2 | 0.5 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.2 | 0.5 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.2 | 4.5 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.2 | 1.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 1.6 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.2 | 0.9 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.2 | 0.8 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.2 | 2.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 3.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 0.8 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 1.6 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.7 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.1 | 1.6 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.3 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 1.0 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.4 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.1 | 1.5 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.9 | GO:1904800 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 | 0.6 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.6 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.1 | 1.6 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 0.1 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 0.7 | GO:2000364 | cardiac muscle tissue regeneration(GO:0061026) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.1 | 2.8 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.4 | GO:2000820 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.1 | 1.2 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.5 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) sensory processing(GO:0050893) |
| 0.1 | 0.9 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 0.7 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 1.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 8.0 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 1.9 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 0.9 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.1 | 2.2 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.1 | 1.5 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 1.0 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.1 | 4.0 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.9 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 4.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 2.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.2 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.4 | GO:0061356 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.6 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 2.3 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 0.6 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 1.0 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 1.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.8 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.1 | 0.7 | GO:0015853 | adenine transport(GO:0015853) |
| 0.1 | 0.6 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.1 | GO:0021896 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.1 | 2.0 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.7 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.1 | 10.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:0043000 | Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.1 | 0.2 | GO:2000553 | positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.1 | 0.9 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.1 | 0.2 | GO:0071264 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.1 | 0.5 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.1 | 6.4 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 1.3 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 4.7 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.9 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 1.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.3 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.1 | 0.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 1.0 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.1 | 1.9 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.9 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.4 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 1.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.4 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.1 | 0.4 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.5 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 2.6 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.1 | 1.7 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.6 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 1.6 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.8 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 1.0 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.3 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.5 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 1.9 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 0.1 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.1 | 0.4 | GO:1903060 | regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.1 | 0.3 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 1.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.9 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.8 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 1.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.3 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 0.4 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.3 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.1 | 0.9 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.1 | 1.9 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.2 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.1 | 0.5 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 0.3 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.1 | 0.3 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.1 | 4.2 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 1.3 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.4 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.1 | 0.1 | GO:0060352 | cell adhesion molecule production(GO:0060352) regulation of cell adhesion molecule production(GO:0060353) positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 0.8 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.3 | GO:1903939 | regulation of TORC2 signaling(GO:1903939) |
| 0.1 | 0.4 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 0.2 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 0.3 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.3 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.1 | 0.5 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 0.3 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 1.0 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.4 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.3 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 1.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 1.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 1.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 6.8 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.1 | 2.8 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 0.2 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.1 | 1.8 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.1 | 0.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.3 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.1 | 0.8 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 0.3 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.1 | 0.2 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.1 | 1.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 1.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.2 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.1 | 0.5 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 1.6 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.1 | 3.6 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 0.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.3 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.5 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 1.8 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.5 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.1 | 1.0 | GO:0008306 | associative learning(GO:0008306) |
| 0.1 | 0.5 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.6 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 1.6 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.1 | 0.4 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.4 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.6 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 0.7 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.4 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 1.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 5.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 12.6 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.1 | 0.7 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.1 | 0.3 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.9 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.1 | 2.5 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 0.4 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 8.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.4 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 1.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.8 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 0.7 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.1 | 0.9 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 0.4 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.7 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 1.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 1.3 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 0.4 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.1 | 0.9 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 2.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.0 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.1 | 0.6 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 5.6 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.1 | 1.7 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 3.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 0.3 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 0.5 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.9 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.1 | 0.8 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 1.1 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.1 | 0.2 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 0.9 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.1 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 0.3 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.2 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.1 | 1.0 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.1 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 0.8 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 1.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 1.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.4 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.8 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.1 | GO:2000744 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.6 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.2 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 1.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.2 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 1.4 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 0.5 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 0.3 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.1 | 0.1 | GO:0099637 | neurotransmitter receptor transport(GO:0099637) |
| 0.1 | 0.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.3 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 | 0.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.2 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.1 | GO:0051532 | regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.1 | 1.8 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 1.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.4 | GO:0002249 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.1 | 3.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.4 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.6 | GO:0010454 | negative regulation of cell fate commitment(GO:0010454) |
| 0.1 | 1.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.1 | GO:0000964 | mitochondrial RNA 5'-end processing(GO:0000964) |
| 0.1 | 0.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.2 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.6 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.1 | 0.3 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.2 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.1 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.2 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.1 | 0.3 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 0.7 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.2 | GO:2000569 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.1 | 0.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.4 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 | 0.2 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.1 | 1.6 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.2 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.1 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.2 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 0.3 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.4 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.1 | 0.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 2.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 0.3 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.8 | GO:2000828 | post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.3 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.3 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 2.0 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 0.8 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.6 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.5 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.5 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.5 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.2 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.1 | GO:2001286 | cellular response to progesterone stimulus(GO:0071393) regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.2 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.4 | GO:0033127 | regulation of histone phosphorylation(GO:0033127) |
| 0.0 | 0.4 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.2 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 5.5 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.2 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.0 | 1.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.7 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.3 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.2 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.6 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 7.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.0 | 0.5 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.2 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.7 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.5 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.0 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.3 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.5 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.2 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.0 | 0.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.2 | GO:0061085 | regulation of histone H3-K27 methylation(GO:0061085) |
| 0.0 | 0.3 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.4 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.3 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0034125 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) |
| 0.0 | 0.9 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.0 | 0.2 | GO:0072144 | glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 0.0 | 0.3 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.0 | 0.3 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.4 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.4 | GO:0010572 | positive regulation of platelet activation(GO:0010572) protein secretion by platelet(GO:0070560) positive regulation of platelet aggregation(GO:1901731) |
| 0.0 | 0.2 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.7 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 1.3 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 2.1 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.9 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.6 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.5 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.4 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.7 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.6 | GO:0070977 | bone maturation(GO:0070977) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0051836 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.3 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.7 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 1.2 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 0.1 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.8 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 8.9 | GO:0097485 | neuron projection guidance(GO:0097485) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.1 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) negative regulation of protein folding(GO:1903333) |
| 0.0 | 1.7 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0050993 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0033034 | positive regulation of myeloid cell apoptotic process(GO:0033034) |
| 0.0 | 0.0 | GO:0002465 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) |
| 0.0 | 0.5 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 8.0 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.4 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.1 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 0.0 | 0.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:0002329 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.5 | GO:0040023 | establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.2 | GO:2000809 | presynaptic membrane organization(GO:0097090) synaptic vesicle clustering(GO:0097091) regulation of presynaptic membrane organization(GO:1901629) positive regulation of presynaptic membrane organization(GO:1901631) regulation of synaptic vesicle clustering(GO:2000807) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 3.5 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.3 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.0 | 0.3 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.8 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.3 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.5 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 0.1 | GO:0034241 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.1 | GO:0036323 | vascular endothelial growth factor receptor-1 signaling pathway(GO:0036323) |
| 0.0 | 0.1 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.4 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.3 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.8 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.4 | GO:0051058 | negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.2 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.1 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.0 | 1.0 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 1.3 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.5 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0033168 | conversion of ds siRNA to ss siRNA involved in RNA interference(GO:0033168) conversion of ds siRNA to ss siRNA(GO:0036404) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.2 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.0 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.0 | GO:1901859 | regulation of mitochondrial DNA replication(GO:0090296) negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.0 | 0.1 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.0 | 0.1 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0002357 | defense response to tumor cell(GO:0002357) |
| 0.0 | 0.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.3 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.5 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.4 | GO:0014075 | response to amine(GO:0014075) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.2 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.4 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.8 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0016129 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.2 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.0 | GO:2001225 | regulation of chloride transport(GO:2001225) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0070203 | regulation of establishment of protein localization to telomere(GO:0070203) regulation of protein localization to chromosome, telomeric region(GO:1904814) positive regulation of protein localization to chromosome, telomeric region(GO:1904816) positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 0.0 | 0.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 18.9 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 1.1 | 5.7 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 1.0 | 7.3 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 1.0 | 5.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.0 | 5.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 1.0 | 1.0 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.9 | 4.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.8 | 14.7 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.6 | 5.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.6 | 26.2 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.6 | 2.9 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.6 | 6.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.5 | 7.1 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.5 | 3.9 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.5 | 3.3 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.5 | 1.8 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.5 | 3.2 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.4 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 8.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.4 | 1.9 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.3 | 6.4 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.3 | 3.6 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.3 | 3.9 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.3 | 3.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.3 | 1.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.3 | 11.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.3 | 5.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 1.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.3 | 1.0 | GO:0071920 | cleavage body(GO:0071920) |
| 0.2 | 1.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 1.3 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.2 | 3.5 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.2 | 1.0 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 0.6 | GO:1905103 | integral component of lysosomal membrane(GO:1905103) |
| 0.2 | 2.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 23.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 1.3 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 0.7 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 0.5 | GO:1990617 | CHOP-ATF4 complex(GO:1990617) |
| 0.2 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.2 | 0.7 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.2 | 1.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.2 | 2.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 8.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 19.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 1.9 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.2 | 0.6 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.2 | 1.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 4.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 1.7 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.2 | 0.8 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.7 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 5.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 13.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.6 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 1.7 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 7.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.7 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 5.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.5 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 2.4 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.1 | 53.9 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 0.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 2.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 15.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.4 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 1.9 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 1.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.3 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 6.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.8 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 2.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 13.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 1.5 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.1 | 0.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 1.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.3 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.9 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 3.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 17.3 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 0.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 3.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.9 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.8 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 2.9 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.6 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.3 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 0.4 | GO:0044423 | virion(GO:0019012) virion part(GO:0044423) |
| 0.1 | 1.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 2.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.2 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 1.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 1.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 2.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.3 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 16.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.3 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 0.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.3 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.4 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 7.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 4.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 10.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 15.2 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 1.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 2.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 4.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 8.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0030849 | autosome(GO:0030849) |
| 0.0 | 1.0 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.4 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 3.0 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 1.0 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 0.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 16.7 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 14.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.9 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.1 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.6 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 1.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 1.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 2.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.3 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.4 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.0 | 0.9 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 2.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 6.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 6.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 1.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 13.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 1.4 | 7.0 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 1.3 | 5.2 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 1.3 | 3.8 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 1.2 | 4.7 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 1.1 | 5.7 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 1.1 | 10.5 | GO:0042835 | BRE binding(GO:0042835) |
| 1.0 | 18.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 1.0 | 8.7 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 1.0 | 2.9 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 1.0 | 2.9 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.9 | 7.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.9 | 5.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.9 | 5.2 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.7 | 11.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.7 | 10.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.7 | 9.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.7 | 19.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.7 | 1.4 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.7 | 2.7 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.6 | 1.9 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.6 | 1.9 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.6 | 0.6 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.6 | 18.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.5 | 2.7 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.5 | 5.8 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.5 | 1.6 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.5 | 2.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.5 | 2.4 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.5 | 8.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.5 | 3.2 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.4 | 5.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.4 | 1.3 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.4 | 14.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.4 | 1.2 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 0.4 | 1.6 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.4 | 1.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.4 | 1.8 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.4 | 2.5 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.3 | 1.0 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.3 | 4.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 1.9 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.3 | 5.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.3 | 1.2 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.3 | 3.9 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.3 | 0.9 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.3 | 1.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 0.9 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.3 | 1.5 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.3 | 1.2 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.3 | 2.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 0.8 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.3 | 3.6 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.3 | 17.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 2.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.3 | 2.6 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.3 | 1.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.3 | 1.3 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.3 | 0.8 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.3 | 1.5 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.3 | 1.5 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 2.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 1.0 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.2 | 2.5 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.2 | 1.7 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.2 | 12.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 0.7 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.2 | 0.7 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 0.2 | 0.2 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.2 | 3.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 0.9 | GO:0009931 | calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 0.7 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.2 | 0.9 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.2 | 0.7 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.2 | 2.2 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.2 | 2.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 0.9 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.2 | 1.1 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.2 | 0.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 2.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 1.9 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 0.8 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.2 | 6.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 1.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 1.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 0.6 | GO:0004314 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) |
| 0.2 | 0.6 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 0.4 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) norepinephrine binding(GO:0051380) |
| 0.2 | 1.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 1.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 0.9 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 1.8 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 0.9 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.2 | 1.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.2 | 0.9 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 0.4 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.2 | 0.5 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.2 | 0.9 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.2 | 3.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 0.5 | GO:0052831 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.2 | 0.7 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.2 | 0.6 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.2 | 0.5 | GO:0004470 | malic enzyme activity(GO:0004470) diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.2 | 1.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 1.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 0.6 | GO:0004513 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.2 | 6.8 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 0.6 | GO:0004420 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.2 | 0.9 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 2.8 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.2 | 5.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 2.6 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 0.6 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 1.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 3.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 2.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.0 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 1.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 1.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.6 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 2.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.4 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 4.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 11.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 3.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 8.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.7 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.3 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.1 | 1.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 1.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 0.4 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 1.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.7 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 1.9 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 1.6 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 1.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 1.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.8 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.1 | 0.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.4 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 0.5 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.1 | 0.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 4.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.4 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.4 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.1 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 2.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.4 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.9 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.1 | 1.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.8 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 0.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.5 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 0.7 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.4 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 0.5 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.1 | 0.8 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.4 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 1.1 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.1 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.3 | GO:0034512 | box C/D snoRNA binding(GO:0034512) |
| 0.1 | 0.4 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.4 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 1.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 2.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.4 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 1.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 1.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 1.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.8 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.3 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 4.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.8 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.5 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.4 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.2 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.1 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 0.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.3 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 1.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.3 | GO:0004639 | phosphoribosylaminoimidazole carboxylase activity(GO:0004638) phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 11.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.1 | 2.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.3 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.1 | 0.4 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.1 | 1.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.0 | GO:0005372 | water transmembrane transporter activity(GO:0005372) |
| 0.1 | 0.9 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 2.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 3.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.3 | GO:0070362 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.1 | 0.3 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 4.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.2 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.4 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.5 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.3 | GO:0004948 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.1 | 1.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.4 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.1 | 1.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 0.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.8 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.1 | 0.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 2.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.7 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 1.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.3 | GO:0030379 | neurotensin receptor activity, non-G-protein coupled(GO:0030379) |
| 0.1 | 0.8 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.3 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 8.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.4 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 0.3 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.5 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 5.7 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 12.9 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.1 | 0.3 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 2.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.4 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.1 | 1.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 3.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.3 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 1.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.6 | GO:0001164 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.3 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.4 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 3.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.1 | GO:0080130 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.3 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 1.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.7 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 0.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.4 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.1 | 0.8 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.2 | GO:0052816 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.1 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.4 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 1.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.3 | GO:0052811 | 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.4 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 2.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 2.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.3 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.5 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.2 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.3 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.3 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.6 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 0.2 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.1 | 3.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 0.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.4 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 1.6 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 3.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0070975 | FHA domain binding(GO:0070975) |
| 0.0 | 4.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 3.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.8 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 5.3 | GO:0043492 | ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 1.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.0 | 0.4 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.4 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 1.2 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.3 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 1.1 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 2.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 8.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.6 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.4 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 14.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 3.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0004615 | phosphomannomutase activity(GO:0004615) |
| 0.0 | 1.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 13.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.4 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 3.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.0 | GO:0022832 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0008513 | secondary active organic cation transmembrane transporter activity(GO:0008513) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.4 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 4.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 0.2 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 1.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.9 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 2.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.0 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0036327 | VEGF-A-activated receptor activity(GO:0036326) VEGF-B-activated receptor activity(GO:0036327) placental growth factor-activated receptor activity(GO:0036332) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 8.5 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 1.0 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.5 | GO:0005253 | anion channel activity(GO:0005253) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 1.8 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 11.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 15.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.3 | 18.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 12.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 10.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 6.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 9.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 5.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 5.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 11.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 4.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 6.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 5.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 2.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 0.6 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 2.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 0.4 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.1 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 0.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 0.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 3.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 3.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 3.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 2.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 2.3 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 8.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 39.0 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.7 | 21.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.6 | 10.0 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.5 | 22.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 0.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.4 | 10.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.4 | 13.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 9.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 5.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 9.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 0.3 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.2 | 2.6 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.2 | 17.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 6.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 1.3 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.2 | 14.9 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.2 | 5.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 5.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 0.7 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 3.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 3.9 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.1 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 3.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 0.5 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 4.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 5.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 8.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 3.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 2.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 4.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 1.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 5.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 0.9 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 5.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 9.6 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.1 | 7.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 3.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.0 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 3.8 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 0.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 2.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 4.7 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 3.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 2.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.9 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.6 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 4.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |