Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for ALX1_ARX
Z-value: 0.62
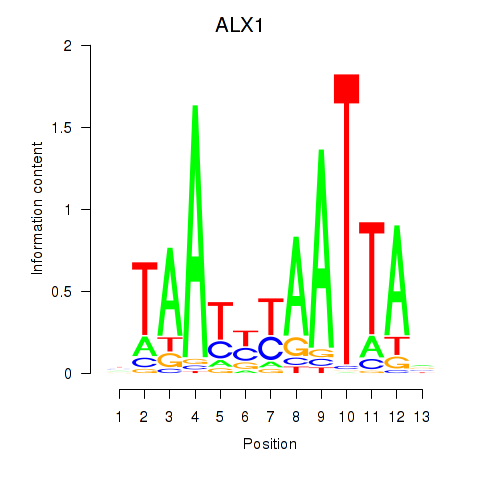
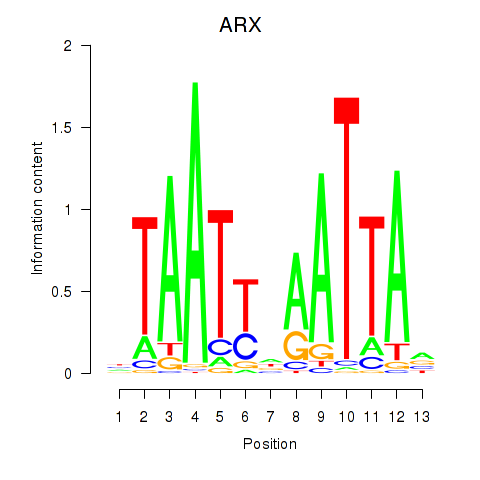
Transcription factors associated with ALX1_ARX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ALX1
|
ENSG00000180318.4 | ALX homeobox 1 |
|
ARX
|
ENSG00000004848.8 | aristaless related homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARX | hg38_v1_chrX_-_25015924_25016002 | -0.24 | 1.8e-01 | Click! |
| ALX1 | hg38_v1_chr12_+_85280199_85280237 | -0.02 | 9.2e-01 | Click! |
Activity profile of ALX1_ARX motif
Sorted Z-values of ALX1_ARX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_195583931 | 4.85 |
ENST00000343267.8
ENST00000421243.5 ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr14_-_94770102 | 3.39 |
ENST00000238558.5
|
GSC
|
goosecoid homeobox |
| chr2_-_218010202 | 3.30 |
ENST00000646520.1
|
TNS1
|
tensin 1 |
| chr10_-_29634964 | 3.17 |
ENST00000375398.6
ENST00000355867.8 |
SVIL
|
supervillin |
| chr1_-_151175966 | 2.71 |
ENST00000441701.1
ENST00000295314.9 |
TMOD4
|
tropomodulin 4 |
| chr6_+_121437378 | 2.38 |
ENST00000650427.1
ENST00000647564.1 |
GJA1
|
gap junction protein alpha 1 |
| chr1_-_91906280 | 2.28 |
ENST00000370399.6
|
TGFBR3
|
transforming growth factor beta receptor 3 |
| chr1_+_170663134 | 2.23 |
ENST00000367760.7
|
PRRX1
|
paired related homeobox 1 |
| chr1_-_178869272 | 2.21 |
ENST00000444255.1
|
ANGPTL1
|
angiopoietin like 1 |
| chr1_-_178871022 | 2.00 |
ENST00000367629.1
|
ANGPTL1
|
angiopoietin like 1 |
| chr1_-_178871060 | 1.91 |
ENST00000234816.7
|
ANGPTL1
|
angiopoietin like 1 |
| chr12_-_91153149 | 1.72 |
ENST00000550758.1
|
DCN
|
decorin |
| chr1_-_93681829 | 1.58 |
ENST00000260502.11
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr10_+_68106109 | 1.49 |
ENST00000540630.5
ENST00000354393.6 |
MYPN
|
myopalladin |
| chr3_-_158106408 | 1.44 |
ENST00000483851.7
|
SHOX2
|
short stature homeobox 2 |
| chr2_+_176129680 | 1.40 |
ENST00000429017.2
ENST00000313173.6 |
HOXD8
|
homeobox D8 |
| chr6_-_65707214 | 1.36 |
ENST00000370621.7
ENST00000393380.6 ENST00000503581.6 |
EYS
|
eyes shut homolog |
| chr14_+_22163226 | 1.35 |
ENST00000390458.3
|
TRAV29DV5
|
T cell receptor alpha variable 29/delta variable 5 |
| chr10_-_49188380 | 1.16 |
ENST00000374153.7
ENST00000374148.1 ENST00000374151.7 |
TMEM273
|
transmembrane protein 273 |
| chr6_-_75363003 | 1.15 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr8_-_6563409 | 1.12 |
ENST00000325203.9
|
ANGPT2
|
angiopoietin 2 |
| chr12_+_12725897 | 1.06 |
ENST00000326765.10
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr3_+_138621207 | 1.02 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr12_-_21775581 | 0.99 |
ENST00000537950.1
ENST00000665145.1 |
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8 |
| chrX_-_11265975 | 0.95 |
ENST00000303025.10
ENST00000657361.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr6_+_34236865 | 0.94 |
ENST00000674029.1
ENST00000447654.5 ENST00000347617.10 ENST00000401473.7 ENST00000311487.9 |
HMGA1
|
high mobility group AT-hook 1 |
| chr7_-_111392915 | 0.90 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr2_+_36696790 | 0.88 |
ENST00000497382.5
ENST00000404084.5 ENST00000379241.7 ENST00000401530.5 |
VIT
|
vitrin |
| chr2_+_36696686 | 0.86 |
ENST00000379242.7
ENST00000389975.7 |
VIT
|
vitrin |
| chr6_-_169250825 | 0.86 |
ENST00000676869.1
ENST00000676760.1 |
THBS2
|
thrombospondin 2 |
| chr11_-_96343170 | 0.84 |
ENST00000524717.6
|
MAML2
|
mastermind like transcriptional coactivator 2 |
| chr3_+_138621225 | 0.84 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr12_+_54016879 | 0.83 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr1_+_186828941 | 0.83 |
ENST00000367466.4
|
PLA2G4A
|
phospholipase A2 group IVA |
| chr19_+_49513353 | 0.82 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr12_-_15662692 | 0.82 |
ENST00000540613.5
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr5_+_40909490 | 0.80 |
ENST00000313164.10
|
C7
|
complement C7 |
| chr12_+_14419136 | 0.79 |
ENST00000545769.1
ENST00000428217.6 ENST00000396279.2 ENST00000542514.5 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr21_+_46635595 | 0.79 |
ENST00000451211.6
ENST00000458387.6 ENST00000397638.6 ENST00000291705.11 ENST00000397637.5 ENST00000334494.8 ENST00000397628.5 ENST00000355680.8 ENST00000440086.5 |
PRMT2
|
protein arginine methyltransferase 2 |
| chr6_-_32941018 | 0.77 |
ENST00000418107.3
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr14_-_21098848 | 0.77 |
ENST00000556174.5
ENST00000554478.5 ENST00000553980.1 ENST00000421093.6 |
ZNF219
|
zinc finger protein 219 |
| chr3_+_12287859 | 0.74 |
ENST00000309576.11
ENST00000397015.7 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr18_-_24311495 | 0.73 |
ENST00000357041.8
|
OSBPL1A
|
oxysterol binding protein like 1A |
| chr1_+_207325629 | 0.73 |
ENST00000618707.2
|
CD55
|
CD55 molecule (Cromer blood group) |
| chr9_-_92536067 | 0.72 |
ENST00000444490.6
|
ECM2
|
extracellular matrix protein 2 |
| chr3_+_12287899 | 0.72 |
ENST00000643888.2
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr5_+_137867868 | 0.71 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chr3_-_125055987 | 0.70 |
ENST00000311127.9
|
HEG1
|
heart development protein with EGF like domains 1 |
| chr5_-_135399863 | 0.68 |
ENST00000510038.1
ENST00000304332.8 |
MACROH2A1
|
macroH2A.1 histone |
| chr9_-_92536031 | 0.66 |
ENST00000344604.9
ENST00000375540.5 |
ECM2
|
extracellular matrix protein 2 |
| chr3_+_69936583 | 0.66 |
ENST00000314557.10
ENST00000394351.9 |
MITF
|
melanocyte inducing transcription factor |
| chr2_+_36696758 | 0.65 |
ENST00000457137.6
|
VIT
|
vitrin |
| chr12_-_118359639 | 0.65 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr8_-_61689768 | 0.61 |
ENST00000517847.6
ENST00000389204.8 ENST00000517661.5 ENST00000517903.5 ENST00000522603.5 ENST00000541428.5 ENST00000522349.5 ENST00000522835.5 ENST00000518306.5 |
ASPH
|
aspartate beta-hydroxylase |
| chr16_+_68085420 | 0.60 |
ENST00000349223.9
|
NFATC3
|
nuclear factor of activated T cells 3 |
| chr17_+_46511511 | 0.59 |
ENST00000576629.5
|
LRRC37A2
|
leucine rich repeat containing 37 member A2 |
| chr16_+_68085552 | 0.58 |
ENST00000329524.8
|
NFATC3
|
nuclear factor of activated T cells 3 |
| chr9_-_92536645 | 0.58 |
ENST00000395534.2
|
ECM2
|
extracellular matrix protein 2 |
| chr18_-_55322334 | 0.57 |
ENST00000630720.3
|
TCF4
|
transcription factor 4 |
| chr3_+_151873634 | 0.56 |
ENST00000362032.6
|
SUCNR1
|
succinate receptor 1 |
| chr3_-_27722316 | 0.55 |
ENST00000449599.4
|
EOMES
|
eomesodermin |
| chr3_-_3109980 | 0.55 |
ENST00000256452.7
ENST00000311981.12 ENST00000430514.6 ENST00000456302.5 |
IL5RA
|
interleukin 5 receptor subunit alpha |
| chr8_-_121641424 | 0.54 |
ENST00000303924.5
|
HAS2
|
hyaluronan synthase 2 |
| chr16_+_68085344 | 0.54 |
ENST00000575270.5
ENST00000346183.8 |
NFATC3
|
nuclear factor of activated T cells 3 |
| chr1_-_79188467 | 0.53 |
ENST00000656300.1
|
ADGRL4
|
adhesion G protein-coupled receptor L4 |
| chr2_+_118942188 | 0.53 |
ENST00000327097.5
|
MARCO
|
macrophage receptor with collagenous structure |
| chr21_-_41926680 | 0.53 |
ENST00000329623.11
|
C2CD2
|
C2 calcium dependent domain containing 2 |
| chr6_-_110358313 | 0.53 |
ENST00000338882.5
|
METTL24
|
methyltransferase like 24 |
| chr2_+_54456311 | 0.52 |
ENST00000615901.4
ENST00000356805.9 |
SPTBN1
|
spectrin beta, non-erythrocytic 1 |
| chr10_-_49188312 | 0.52 |
ENST00000453436.5
ENST00000474718.5 |
TMEM273
|
transmembrane protein 273 |
| chr18_-_55321986 | 0.51 |
ENST00000570287.6
|
TCF4
|
transcription factor 4 |
| chr6_+_54099565 | 0.51 |
ENST00000511678.5
|
MLIP
|
muscular LMNA interacting protein |
| chr4_+_94455245 | 0.50 |
ENST00000508216.5
ENST00000514743.5 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr4_-_67963441 | 0.50 |
ENST00000508048.6
|
TMPRSS11A
|
transmembrane serine protease 11A |
| chr11_-_16397521 | 0.49 |
ENST00000533411.5
|
SOX6
|
SRY-box transcription factor 6 |
| chr2_-_88861920 | 0.49 |
ENST00000390242.2
|
IGKJ1
|
immunoglobulin kappa joining 1 |
| chrX_-_15314543 | 0.49 |
ENST00000344384.8
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr19_-_43465596 | 0.46 |
ENST00000244333.4
|
LYPD3
|
LY6/PLAUR domain containing 3 |
| chr16_-_21431553 | 0.46 |
ENST00000534903.1
|
NPIPB3
|
nuclear pore complex interacting protein family member B3 |
| chr12_-_89352395 | 0.46 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr1_+_159008978 | 0.46 |
ENST00000447473.6
|
IFI16
|
interferon gamma inducible protein 16 |
| chr15_+_100877714 | 0.45 |
ENST00000561338.5
|
ALDH1A3
|
aldehyde dehydrogenase 1 family member A3 |
| chr22_-_28306645 | 0.44 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr3_+_69936629 | 0.44 |
ENST00000394348.2
ENST00000531774.1 |
MITF
|
melanocyte inducing transcription factor |
| chr4_+_68447453 | 0.43 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr18_-_55322215 | 0.42 |
ENST00000457482.7
|
TCF4
|
transcription factor 4 |
| chrX_-_15600953 | 0.42 |
ENST00000679212.1
ENST00000679278.1 ENST00000678046.1 ENST00000252519.8 |
ACE2
|
angiotensin I converting enzyme 2 |
| chr7_-_93148345 | 0.41 |
ENST00000437805.5
ENST00000446959.5 ENST00000439952.5 ENST00000414791.5 ENST00000446033.1 ENST00000411955.5 ENST00000318238.9 |
SAMD9L
|
sterile alpha motif domain containing 9 like |
| chr3_-_196515315 | 0.40 |
ENST00000397537.3
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chrX_-_19799751 | 0.39 |
ENST00000379698.8
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chr12_-_89352487 | 0.38 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr15_+_41774459 | 0.38 |
ENST00000457542.7
ENST00000456763.6 |
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr1_-_79188390 | 0.37 |
ENST00000662530.1
|
ADGRL4
|
adhesion G protein-coupled receptor L4 |
| chr8_+_85987287 | 0.36 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase H+ transporting V0 subunit d2 |
| chr2_+_161136901 | 0.35 |
ENST00000259075.6
ENST00000432002.5 |
TANK
|
TRAF family member associated NFKB activator |
| chrX_+_108044967 | 0.35 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr6_+_54099538 | 0.34 |
ENST00000447836.6
|
MLIP
|
muscular LMNA interacting protein |
| chr5_+_141364153 | 0.32 |
ENST00000518069.2
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr1_+_24556087 | 0.31 |
ENST00000374392.3
|
NCMAP
|
non-compact myelin associated protein |
| chr12_+_57610150 | 0.31 |
ENST00000333972.11
|
ARHGEF25
|
Rho guanine nucleotide exchange factor 25 |
| chr6_-_132659178 | 0.30 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr16_-_21864103 | 0.29 |
ENST00000541674.5
|
NPIPB4
|
nuclear pore complex interacting protein family member B4 |
| chr8_-_85341659 | 0.29 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr2_+_28778848 | 0.29 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1 catalytic subunit beta |
| chrX_-_55182442 | 0.29 |
ENST00000545075.3
|
MTRNR2L10
|
MT-RNR2 like 10 |
| chr22_-_30246739 | 0.28 |
ENST00000403987.3
ENST00000249075.4 |
LIF
|
LIF interleukin 6 family cytokine |
| chr10_-_101229449 | 0.28 |
ENST00000370193.4
|
LBX1
|
ladybird homeobox 1 |
| chr2_+_118942290 | 0.28 |
ENST00000412481.1
|
MARCO
|
macrophage receptor with collagenous structure |
| chr11_-_13496018 | 0.27 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr1_-_110391041 | 0.27 |
ENST00000369781.8
ENST00000437429.6 ENST00000541986.5 |
SLC16A4
|
solute carrier family 16 member 4 |
| chr7_-_7535863 | 0.27 |
ENST00000399429.8
|
COL28A1
|
collagen type XXVIII alpha 1 chain |
| chr6_+_3258914 | 0.27 |
ENST00000438998.7
ENST00000419065.6 ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome assembly chaperone 4 |
| chr11_+_66011994 | 0.26 |
ENST00000312134.3
|
CST6
|
cystatin E/M |
| chr12_-_68225806 | 0.26 |
ENST00000229134.5
|
IL26
|
interleukin 26 |
| chr7_-_55552741 | 0.26 |
ENST00000418904.5
|
VOPP1
|
VOPP1 WW domain binding protein |
| chr16_-_3372666 | 0.26 |
ENST00000399974.5
|
MTRNR2L4
|
MT-RNR2 like 4 |
| chr11_-_102705737 | 0.25 |
ENST00000260229.5
|
MMP27
|
matrix metallopeptidase 27 |
| chr8_+_38820332 | 0.25 |
ENST00000518809.5
ENST00000520611.1 |
TACC1
|
transforming acidic coiled-coil containing protein 1 |
| chr12_-_118359105 | 0.25 |
ENST00000541186.5
ENST00000539872.5 |
TAOK3
|
TAO kinase 3 |
| chr16_+_53435612 | 0.25 |
ENST00000544405.6
|
RBL2
|
RB transcriptional corepressor like 2 |
| chr5_+_146447304 | 0.24 |
ENST00000296702.9
ENST00000394421.7 ENST00000679501.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr14_+_23240346 | 0.24 |
ENST00000430154.6
|
RNF212B
|
ring finger protein 212B |
| chr7_+_107583919 | 0.24 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr6_+_112236806 | 0.24 |
ENST00000588837.5
ENST00000590293.5 ENST00000585450.5 ENST00000629766.2 ENST00000590804.5 ENST00000590584.4 ENST00000628122.2 ENST00000627025.1 ENST00000590673.5 ENST00000585611.5 ENST00000587816.2 |
LAMA4-AS1
ENSG00000281613.2
|
LAMA4 antisense RNA 1 novel ret finger protein-like 4B |
| chr8_-_85341705 | 0.23 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr16_+_24537693 | 0.23 |
ENST00000564314.5
ENST00000567686.5 |
RBBP6
|
RB binding protein 6, ubiquitin ligase |
| chr15_-_43800132 | 0.23 |
ENST00000299969.10
ENST00000319327.7 |
SERINC4
|
serine incorporator 4 |
| chrX_+_1591590 | 0.23 |
ENST00000313871.9
ENST00000381261.8 |
AKAP17A
|
A-kinase anchoring protein 17A |
| chr6_+_30914329 | 0.23 |
ENST00000541562.6
|
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr6_+_30914205 | 0.23 |
ENST00000672801.1
ENST00000321897.9 ENST00000625423.2 ENST00000676266.1 ENST00000428017.5 |
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr1_+_186296267 | 0.22 |
ENST00000533951.5
ENST00000367482.8 ENST00000635041.1 ENST00000367483.8 ENST00000367485.4 ENST00000445192.7 |
PRG4
|
proteoglycan 4 |
| chr3_+_130850585 | 0.22 |
ENST00000505330.5
ENST00000504381.5 ENST00000507488.6 |
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr4_-_89029881 | 0.21 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13 member A |
| chr19_-_6393131 | 0.21 |
ENST00000394456.10
|
GTF2F1
|
general transcription factor IIF subunit 1 |
| chr7_+_20615653 | 0.21 |
ENST00000404938.7
|
ABCB5
|
ATP binding cassette subfamily B member 5 |
| chr9_-_135639540 | 0.21 |
ENST00000371763.6
ENST00000613244.1 |
GLT6D1
|
glycosyltransferase 6 domain containing 1 |
| chr1_-_21050952 | 0.21 |
ENST00000264211.12
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma 3 |
| chrX_+_108045050 | 0.21 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr3_+_140941792 | 0.21 |
ENST00000446041.6
ENST00000324194.12 ENST00000507429.5 |
SLC25A36
|
solute carrier family 25 member 36 |
| chr5_-_60488055 | 0.21 |
ENST00000505507.6
ENST00000515835.2 ENST00000502484.6 |
PDE4D
|
phosphodiesterase 4D |
| chr4_-_82848843 | 0.20 |
ENST00000511338.1
|
SEC31A
|
SEC31 homolog A, COPII coat complex component |
| chr11_-_58268260 | 0.20 |
ENST00000395079.2
|
OR10W1
|
olfactory receptor family 10 subfamily W member 1 |
| chr13_+_53028806 | 0.19 |
ENST00000219022.3
|
OLFM4
|
olfactomedin 4 |
| chr9_+_121567057 | 0.19 |
ENST00000394340.7
ENST00000436835.5 ENST00000259371.6 |
DAB2IP
|
DAB2 interacting protein |
| chr10_-_110304894 | 0.19 |
ENST00000369603.10
|
SMNDC1
|
survival motor neuron domain containing 1 |
| chr6_+_3258888 | 0.19 |
ENST00000380305.4
|
PSMG4
|
proteasome assembly chaperone 4 |
| chr6_-_154430495 | 0.19 |
ENST00000424998.3
|
CNKSR3
|
CNKSR family member 3 |
| chr19_-_6393205 | 0.18 |
ENST00000595047.5
|
GTF2F1
|
general transcription factor IIF subunit 1 |
| chr2_-_144520315 | 0.18 |
ENST00000465070.5
ENST00000465308.5 ENST00000636471.1 ENST00000629520.2 ENST00000675069.1 ENST00000636026.2 ENST00000444559.5 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr6_-_111483700 | 0.18 |
ENST00000435970.5
ENST00000358835.7 |
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chrX_-_32412220 | 0.18 |
ENST00000619831.5
|
DMD
|
dystrophin |
| chr18_+_68798065 | 0.18 |
ENST00000360242.9
|
CCDC102B
|
coiled-coil domain containing 102B |
| chr12_+_59689337 | 0.17 |
ENST00000261187.8
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr3_-_27722699 | 0.16 |
ENST00000461503.2
|
EOMES
|
eomesodermin |
| chr15_+_63529142 | 0.16 |
ENST00000268049.11
|
USP3
|
ubiquitin specific peptidase 3 |
| chrX_+_16650155 | 0.16 |
ENST00000380200.3
|
S100G
|
S100 calcium binding protein G |
| chr17_-_41118369 | 0.16 |
ENST00000391413.4
|
KRTAP4-11
|
keratin associated protein 4-11 |
| chr11_-_111876567 | 0.16 |
ENST00000526587.2
|
ENSG00000258529.5
|
novel protein |
| chr4_+_75724569 | 0.16 |
ENST00000514213.7
ENST00000264904.8 |
USO1
|
USO1 vesicle transport factor |
| chr15_-_52678560 | 0.16 |
ENST00000562351.2
ENST00000261844.11 ENST00000399202.8 ENST00000562135.5 |
FAM214A
|
family with sequence similarity 214 member A |
| chr15_-_43106022 | 0.15 |
ENST00000627960.1
ENST00000290650.9 |
UBR1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr12_-_52553139 | 0.15 |
ENST00000267119.6
|
KRT71
|
keratin 71 |
| chr12_-_118190510 | 0.15 |
ENST00000540561.5
ENST00000537952.1 ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr6_+_27824084 | 0.15 |
ENST00000355057.3
|
H4C11
|
H4 clustered histone 11 |
| chr7_+_142384328 | 0.15 |
ENST00000390361.3
|
TRBV7-3
|
T cell receptor beta variable 7-3 |
| chr17_-_62808339 | 0.15 |
ENST00000583600.5
|
MARCHF10
|
membrane associated ring-CH-type finger 10 |
| chr15_+_57219411 | 0.14 |
ENST00000543579.5
ENST00000537840.5 ENST00000343827.7 |
TCF12
|
transcription factor 12 |
| chr4_-_142305935 | 0.14 |
ENST00000511838.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr17_-_62808299 | 0.14 |
ENST00000311269.10
|
MARCHF10
|
membrane associated ring-CH-type finger 10 |
| chr12_-_57479552 | 0.13 |
ENST00000424809.6
|
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr5_+_141364231 | 0.13 |
ENST00000611914.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr17_+_44957907 | 0.13 |
ENST00000678938.1
|
NMT1
|
N-myristoyltransferase 1 |
| chr5_+_141338753 | 0.13 |
ENST00000528330.2
ENST00000394576.3 |
PCDHGA2
|
protocadherin gamma subfamily A, 2 |
| chr5_+_69565122 | 0.13 |
ENST00000507595.1
|
GTF2H2C
|
GTF2H2 family member C |
| chr4_+_185204237 | 0.12 |
ENST00000618785.4
ENST00000504273.5 |
SNX25
|
sorting nexin 25 |
| chr14_+_21098819 | 0.12 |
ENST00000418511.6
ENST00000554329.6 |
TMEM253
|
transmembrane protein 253 |
| chr21_-_30598902 | 0.12 |
ENST00000334897.4
|
KRTAP6-2
|
keratin associated protein 6-2 |
| chrX_-_41665766 | 0.12 |
ENST00000643043.2
ENST00000486402.1 ENST00000646087.2 |
CASK
|
calcium/calmodulin dependent serine protein kinase |
| chr17_-_40799939 | 0.12 |
ENST00000306658.8
|
KRT28
|
keratin 28 |
| chr20_+_31547367 | 0.11 |
ENST00000394552.3
|
MCTS2P
|
MCTS family member 2, pseudogene |
| chr17_-_62808283 | 0.11 |
ENST00000544856.6
|
MARCHF10
|
membrane associated ring-CH-type finger 10 |
| chr6_-_26199272 | 0.11 |
ENST00000650491.1
ENST00000635200.1 ENST00000341023.2 |
ENSG00000282988.2
H2AC7
|
novel protein H2A clustered histone 7 |
| chr3_-_151278359 | 0.11 |
ENST00000494668.1
|
P2RY14
|
purinergic receptor P2Y14 |
| chr8_-_16567362 | 0.11 |
ENST00000518026.5
|
MSR1
|
macrophage scavenger receptor 1 |
| chr3_-_47892743 | 0.11 |
ENST00000420772.6
|
MAP4
|
microtubule associated protein 4 |
| chr12_+_53461015 | 0.11 |
ENST00000553064.6
ENST00000547859.2 |
PCBP2
|
poly(rC) binding protein 2 |
| chrX_+_38352573 | 0.11 |
ENST00000039007.5
|
OTC
|
ornithine transcarbamylase |
| chr4_-_185657520 | 0.11 |
ENST00000438278.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr17_+_73232400 | 0.11 |
ENST00000535032.7
ENST00000577615.5 ENST00000585109.5 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr2_+_43774033 | 0.11 |
ENST00000260605.12
ENST00000406852.7 ENST00000398823.6 ENST00000605786.5 |
DYNC2LI1
|
dynein cytoplasmic 2 light intermediate chain 1 |
| chr4_-_142305826 | 0.10 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr9_+_72577369 | 0.10 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr3_-_165078480 | 0.10 |
ENST00000264382.8
|
SI
|
sucrase-isomaltase |
| chrX_-_77634229 | 0.10 |
ENST00000675732.1
|
ATRX
|
ATRX chromatin remodeler |
| chr4_+_94207596 | 0.10 |
ENST00000359052.8
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr12_-_110499546 | 0.10 |
ENST00000552130.6
|
VPS29
|
VPS29 retromer complex component |
| chr14_+_67364849 | 0.10 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2 subunit alpha |
| chr3_-_12545499 | 0.10 |
ENST00000564146.4
|
MKRN2OS
|
MKRN2 opposite strand |
| chr7_-_116159886 | 0.09 |
ENST00000484212.5
|
TFEC
|
transcription factor EC |
| chr17_-_73232218 | 0.09 |
ENST00000583024.1
ENST00000403627.7 ENST00000405159.7 ENST00000581110.1 |
FAM104A
|
family with sequence similarity 104 member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of ALX1_ARX
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 1.1 | 3.4 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.6 | 2.4 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.5 | 2.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.4 | 1.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.3 | 0.9 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.3 | 0.8 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.3 | 2.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 0.8 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.2 | 0.7 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.2 | 0.7 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.2 | 0.8 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 0.9 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.2 | 1.5 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 0.8 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 2.5 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.4 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.5 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.7 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 1.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.8 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.4 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 0.5 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 1.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.5 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.5 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.6 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.8 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.4 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 0.2 | GO:0048058 | compound eye corneal lens development(GO:0048058) |
| 0.1 | 1.7 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.1 | 0.3 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 0.2 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 1.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 1.4 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 | 1.0 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.3 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 3.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.8 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.4 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 1.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.0 | 1.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:1904908 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.0 | 1.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.0 | 3.3 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 1.0 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0002665 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.9 | GO:2000142 | regulation of DNA-templated transcription, initiation(GO:2000142) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.8 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.2 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.6 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 2.6 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.6 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.5 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.3 | 0.8 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.2 | 0.9 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 3.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.4 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 1.7 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 4.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.9 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 2.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.5 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 2.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0044207 | eukaryotic 48S preinitiation complex(GO:0033290) translation initiation ternary complex(GO:0044207) |
| 0.0 | 0.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 4.4 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 6.3 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.4 | 2.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 2.4 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.3 | 0.8 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 0.8 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 1.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.4 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 1.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.5 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.6 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.4 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 3.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.3 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 3.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 4.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 1.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 1.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 1.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 3.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 1.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.8 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 1.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 5.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 4.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 3.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 6.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 2.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |