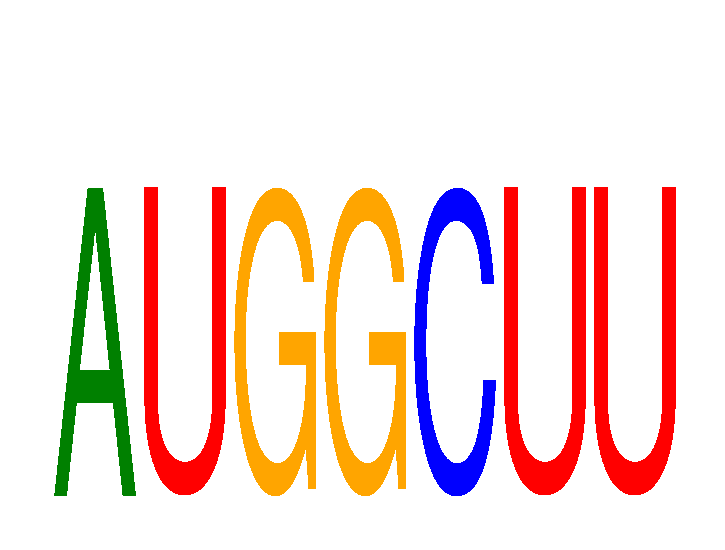Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for AUGGCUU
Z-value: 1.48
miRNA associated with seed AUGGCUU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-135a-5p
|
MIMAT0000428 |
|
hsa-miR-135b-5p
|
MIMAT0000758 |
Activity profile of AUGGCUU motif
Sorted Z-values of AUGGCUU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_175796310 | 9.49 |
ENST00000359546.8
|
CPLX2
|
complexin 2 |
| chr8_+_24914942 | 8.60 |
ENST00000433454.3
|
NEFM
|
neurofilament medium |
| chr9_-_19786928 | 7.46 |
ENST00000341998.6
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 member 2 |
| chr9_-_90642791 | 6.75 |
ENST00000375765.5
ENST00000636786.1 |
DIRAS2
|
DIRAS family GTPase 2 |
| chr19_-_17688326 | 5.95 |
ENST00000552293.5
ENST00000551649.5 ENST00000519716.7 ENST00000550896.1 |
UNC13A
|
unc-13 homolog A |
| chr15_+_26867047 | 5.77 |
ENST00000335625.10
ENST00000555182.5 ENST00000400081.7 |
GABRA5
|
gamma-aminobutyric acid type A receptor subunit alpha5 |
| chr12_+_78864768 | 5.68 |
ENST00000261205.9
ENST00000457153.6 |
SYT1
|
synaptotagmin 1 |
| chr1_+_159171607 | 5.14 |
ENST00000368124.8
ENST00000368125.9 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr12_-_13981544 | 5.13 |
ENST00000609686.4
|
GRIN2B
|
glutamate ionotropic receptor NMDA type subunit 2B |
| chr15_+_91100194 | 5.12 |
ENST00000394232.6
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr11_-_118176576 | 5.05 |
ENST00000278947.6
|
SCN2B
|
sodium voltage-gated channel beta subunit 2 |
| chr1_+_209583706 | 5.02 |
ENST00000361322.3
ENST00000651530.1 ENST00000009105.5 ENST00000423146.5 |
CAMK1G
|
calcium/calmodulin dependent protein kinase IG |
| chr5_-_137499293 | 4.90 |
ENST00000510689.5
ENST00000394945.6 |
SPOCK1
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 1 |
| chr16_+_58463663 | 4.90 |
ENST00000258187.9
|
NDRG4
|
NDRG family member 4 |
| chr3_+_49554436 | 4.78 |
ENST00000296452.5
|
BSN
|
bassoon presynaptic cytomatrix protein |
| chr7_+_31052297 | 4.76 |
ENST00000304166.9
|
ADCYAP1R1
|
ADCYAP receptor type I |
| chr5_-_45696326 | 4.73 |
ENST00000673735.1
ENST00000303230.6 |
HCN1
|
hyperpolarization activated cyclic nucleotide gated potassium channel 1 |
| chrX_-_155264471 | 4.69 |
ENST00000369454.4
|
RAB39B
|
RAB39B, member RAS oncogene family |
| chr18_+_74534594 | 4.59 |
ENST00000582365.1
|
CNDP1
|
carnosine dipeptidase 1 |
| chr6_+_24126186 | 4.57 |
ENST00000378478.5
ENST00000378491.9 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr5_-_161546708 | 4.47 |
ENST00000393959.6
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr6_+_122996227 | 4.42 |
ENST00000275162.10
|
CLVS2
|
clavesin 2 |
| chr9_+_74497308 | 4.37 |
ENST00000376896.8
|
RORB
|
RAR related orphan receptor B |
| chr6_-_99349647 | 4.18 |
ENST00000389677.6
|
FAXC
|
failed axon connections homolog, metaxin like GST domain containing |
| chr19_+_589873 | 4.12 |
ENST00000251287.3
|
HCN2
|
hyperpolarization activated cyclic nucleotide gated potassium and sodium channel 2 |
| chr17_-_10026265 | 4.10 |
ENST00000437099.6
ENST00000396115.6 |
GAS7
|
growth arrest specific 7 |
| chr10_-_1737516 | 3.94 |
ENST00000381312.6
|
ADARB2
|
adenosine deaminase RNA specific B2 (inactive) |
| chr7_+_30134956 | 3.90 |
ENST00000324453.13
ENST00000409688.1 |
MTURN
|
maturin, neural progenitor differentiation regulator homolog |
| chr4_-_826113 | 3.82 |
ENST00000304062.11
|
CPLX1
|
complexin 1 |
| chr6_+_138161932 | 3.71 |
ENST00000251691.5
|
ARFGEF3
|
ARFGEF family member 3 |
| chr4_+_47031551 | 3.61 |
ENST00000295454.8
|
GABRB1
|
gamma-aminobutyric acid type A receptor subunit beta1 |
| chr15_+_59438149 | 3.55 |
ENST00000288228.10
ENST00000559628.5 ENST00000557914.5 ENST00000560474.5 |
FAM81A
|
family with sequence similarity 81 member A |
| chr11_+_62707668 | 3.50 |
ENST00000294117.6
|
GNG3
|
G protein subunit gamma 3 |
| chr2_+_165239388 | 3.48 |
ENST00000424833.5
ENST00000375437.7 ENST00000631182.3 |
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr1_-_229342489 | 3.40 |
ENST00000366687.5
|
CCSAP
|
centriole, cilia and spindle associated protein |
| chr17_-_7929793 | 3.33 |
ENST00000303790.3
|
KCNAB3
|
potassium voltage-gated channel subfamily A regulatory beta subunit 3 |
| chr16_-_705726 | 3.26 |
ENST00000397621.6
ENST00000324361.9 |
FBXL16
|
F-box and leucine rich repeat protein 16 |
| chr2_-_40452046 | 3.23 |
ENST00000406785.6
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr19_-_55443263 | 3.18 |
ENST00000416792.2
ENST00000376325.10 |
SHISA7
|
shisa family member 7 |
| chr20_-_49482645 | 3.14 |
ENST00000371741.6
|
KCNB1
|
potassium voltage-gated channel subfamily B member 1 |
| chrX_-_24672654 | 3.11 |
ENST00000379145.5
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr6_+_96015964 | 2.98 |
ENST00000302103.6
|
FUT9
|
fucosyltransferase 9 |
| chr17_-_80476597 | 2.93 |
ENST00000306773.5
|
NPTX1
|
neuronal pentraxin 1 |
| chr17_+_42682470 | 2.92 |
ENST00000264638.9
|
CNTNAP1
|
contactin associated protein 1 |
| chr1_+_161258738 | 2.88 |
ENST00000504449.2
|
PCP4L1
|
Purkinje cell protein 4 like 1 |
| chr1_+_107141022 | 2.69 |
ENST00000370067.5
ENST00000370068.6 |
NTNG1
|
netrin G1 |
| chr19_-_38224215 | 2.66 |
ENST00000355526.10
ENST00000420980.7 ENST00000614244.4 |
DPF1
|
double PHD fingers 1 |
| chr1_+_162069674 | 2.66 |
ENST00000361897.10
|
NOS1AP
|
nitric oxide synthase 1 adaptor protein |
| chr15_-_48178144 | 2.60 |
ENST00000616409.4
ENST00000324324.12 ENST00000610570.4 |
MYEF2
|
myelin expression factor 2 |
| chrX_+_12138426 | 2.60 |
ENST00000380682.5
ENST00000675598.1 |
FRMPD4
|
FERM and PDZ domain containing 4 |
| chr19_-_2702682 | 2.60 |
ENST00000382159.8
|
GNG7
|
G protein subunit gamma 7 |
| chr2_-_2331225 | 2.56 |
ENST00000648627.1
ENST00000649663.1 ENST00000650560.1 ENST00000428368.7 ENST00000648316.1 ENST00000648665.1 ENST00000649313.1 ENST00000399161.7 ENST00000647738.2 |
MYT1L
|
myelin transcription factor 1 like |
| chr6_+_11093753 | 2.54 |
ENST00000416247.4
|
SMIM13
|
small integral membrane protein 13 |
| chr8_+_103819244 | 2.54 |
ENST00000262231.14
ENST00000507740.5 ENST00000408894.6 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr17_+_58755821 | 2.48 |
ENST00000308249.4
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent 1E |
| chr3_-_56468346 | 2.42 |
ENST00000288221.11
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr9_-_10612966 | 2.38 |
ENST00000381196.9
|
PTPRD
|
protein tyrosine phosphatase receptor type D |
| chr19_-_43639788 | 2.35 |
ENST00000222374.3
|
CADM4
|
cell adhesion molecule 4 |
| chr13_-_67230377 | 2.32 |
ENST00000544246.5
ENST00000377861.4 |
PCDH9
|
protocadherin 9 |
| chr8_-_132481057 | 2.29 |
ENST00000388996.10
|
KCNQ3
|
potassium voltage-gated channel subfamily Q member 3 |
| chr1_+_171841466 | 2.28 |
ENST00000367733.6
ENST00000627582.3 ENST00000355305.9 ENST00000367731.5 |
DNM3
|
dynamin 3 |
| chrX_-_13938618 | 2.21 |
ENST00000454189.6
|
GPM6B
|
glycoprotein M6B |
| chr4_-_86360071 | 2.19 |
ENST00000641677.1
ENST00000639234.1 ENST00000641553.1 ENST00000641826.1 ENST00000641537.1 ENST00000395169.9 ENST00000641408.1 ENST00000638225.1 ENST00000641052.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr8_+_93916882 | 2.17 |
ENST00000297598.5
ENST00000520728.5 ENST00000518107.5 ENST00000396200.3 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr1_+_42380772 | 2.13 |
ENST00000431473.4
ENST00000410070.6 |
RIMKLA
|
ribosomal modification protein rimK like family member A |
| chr1_-_72282457 | 2.06 |
ENST00000357731.10
|
NEGR1
|
neuronal growth regulator 1 |
| chr19_-_50637939 | 2.03 |
ENST00000338916.8
|
SYT3
|
synaptotagmin 3 |
| chr11_-_117316230 | 1.97 |
ENST00000313005.11
ENST00000528053.5 |
BACE1
|
beta-secretase 1 |
| chr18_+_34978244 | 1.95 |
ENST00000436190.6
|
MAPRE2
|
microtubule associated protein RP/EB family member 2 |
| chr7_-_88220025 | 1.94 |
ENST00000419179.5
ENST00000265729.7 |
SRI
|
sorcin |
| chr1_-_233295712 | 1.94 |
ENST00000258229.14
|
PCNX2
|
pecanex 2 |
| chr7_+_141074038 | 1.92 |
ENST00000565468.6
ENST00000610315.1 |
TMEM178B
|
transmembrane protein 178B |
| chrX_+_153517672 | 1.90 |
ENST00000349466.6
ENST00000370186.5 ENST00000359149.8 |
ATP2B3
|
ATPase plasma membrane Ca2+ transporting 3 |
| chr7_-_76358982 | 1.89 |
ENST00000307630.5
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma |
| chr7_-_87059639 | 1.86 |
ENST00000450689.7
|
ELAPOR2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr12_+_51424802 | 1.84 |
ENST00000453097.7
|
SLC4A8
|
solute carrier family 4 member 8 |
| chr3_+_160225409 | 1.83 |
ENST00000326474.5
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr22_-_36703723 | 1.83 |
ENST00000300105.7
|
CACNG2
|
calcium voltage-gated channel auxiliary subunit gamma 2 |
| chr8_+_38176802 | 1.82 |
ENST00000287322.5
|
BAG4
|
BAG cochaperone 4 |
| chr3_+_138347648 | 1.81 |
ENST00000614350.4
ENST00000289104.8 |
MRAS
|
muscle RAS oncogene homolog |
| chr8_-_19013693 | 1.75 |
ENST00000327040.13
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr16_+_8720706 | 1.75 |
ENST00000425191.6
ENST00000569156.5 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr9_-_125143457 | 1.72 |
ENST00000373549.8
ENST00000336505.11 |
SCAI
|
suppressor of cancer cell invasion |
| chr6_-_11044275 | 1.70 |
ENST00000354666.4
|
ELOVL2
|
ELOVL fatty acid elongase 2 |
| chr7_-_32299287 | 1.70 |
ENST00000396193.5
|
PDE1C
|
phosphodiesterase 1C |
| chr10_-_60389833 | 1.69 |
ENST00000280772.7
|
ANK3
|
ankyrin 3 |
| chr1_-_37034492 | 1.69 |
ENST00000373091.8
|
GRIK3
|
glutamate ionotropic receptor kainate type subunit 3 |
| chr6_+_17393576 | 1.69 |
ENST00000229922.7
ENST00000611958.4 |
CAP2
|
cyclase associated actin cytoskeleton regulatory protein 2 |
| chr11_-_119729158 | 1.69 |
ENST00000264025.8
|
NECTIN1
|
nectin cell adhesion molecule 1 |
| chr4_+_98261368 | 1.69 |
ENST00000509011.5
ENST00000408927.8 ENST00000380158.8 ENST00000264572.11 |
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr9_-_76906090 | 1.68 |
ENST00000376718.8
|
PRUNE2
|
prune homolog 2 with BCH domain |
| chr14_-_54489003 | 1.68 |
ENST00000554908.5
ENST00000616146.4 |
GMFB
|
glia maturation factor beta |
| chr12_+_100794769 | 1.66 |
ENST00000392977.8
ENST00000546991.1 ENST00000392979.7 |
ANO4
|
anoctamin 4 |
| chr7_+_152759744 | 1.65 |
ENST00000377776.7
ENST00000256001.13 ENST00000397282.2 |
ACTR3B
|
actin related protein 3B |
| chr11_-_13463168 | 1.64 |
ENST00000526841.1
ENST00000278174.10 ENST00000529708.5 ENST00000528120.5 |
BTBD10
|
BTB domain containing 10 |
| chr17_-_8163522 | 1.63 |
ENST00000404970.3
|
VAMP2
|
vesicle associated membrane protein 2 |
| chr4_-_42657085 | 1.63 |
ENST00000264449.14
ENST00000510289.1 ENST00000381668.9 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr5_-_147782518 | 1.61 |
ENST00000507386.5
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr10_+_115093331 | 1.56 |
ENST00000609571.5
ENST00000355044.8 ENST00000526946.5 |
ATRNL1
|
attractin like 1 |
| chr1_-_100894775 | 1.56 |
ENST00000416479.1
ENST00000370113.7 |
EXTL2
|
exostosin like glycosyltransferase 2 |
| chr11_+_121452291 | 1.54 |
ENST00000260197.12
|
SORL1
|
sortilin related receptor 1 |
| chr12_+_108131740 | 1.52 |
ENST00000332082.8
|
WSCD2
|
WSC domain containing 2 |
| chr2_-_174487005 | 1.49 |
ENST00000392552.7
ENST00000295500.8 ENST00000614352.4 ENST00000392551.6 |
GPR155
|
G protein-coupled receptor 155 |
| chr21_-_37916440 | 1.45 |
ENST00000609713.2
|
KCNJ6
|
potassium inwardly rectifying channel subfamily J member 6 |
| chr20_-_9838831 | 1.40 |
ENST00000378423.5
|
PAK5
|
p21 (RAC1) activated kinase 5 |
| chr20_-_33443651 | 1.39 |
ENST00000217381.3
|
SNTA1
|
syntrophin alpha 1 |
| chr18_+_57352541 | 1.39 |
ENST00000324000.4
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr1_+_109984756 | 1.36 |
ENST00000393614.8
ENST00000369799.10 |
AHCYL1
|
adenosylhomocysteinase like 1 |
| chr18_+_905103 | 1.36 |
ENST00000579794.1
|
ADCYAP1
|
adenylate cyclase activating polypeptide 1 |
| chr2_-_179861604 | 1.27 |
ENST00000410066.7
|
ZNF385B
|
zinc finger protein 385B |
| chr10_+_103493931 | 1.25 |
ENST00000369780.8
|
NEURL1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr19_+_18153155 | 1.24 |
ENST00000222254.13
|
PIK3R2
|
phosphoinositide-3-kinase regulatory subunit 2 |
| chr13_+_97434154 | 1.24 |
ENST00000245304.5
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr9_-_71121596 | 1.23 |
ENST00000377110.9
ENST00000377111.8 ENST00000677713.2 |
TRPM3
|
transient receptor potential cation channel subfamily M member 3 |
| chr10_+_101354083 | 1.23 |
ENST00000408038.6
ENST00000370187.8 |
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr7_-_117873420 | 1.23 |
ENST00000160373.8
|
CTTNBP2
|
cortactin binding protein 2 |
| chr8_-_20303955 | 1.17 |
ENST00000381569.5
|
LZTS1
|
leucine zipper tumor suppressor 1 |
| chr20_+_44247298 | 1.14 |
ENST00000342560.10
ENST00000438466.5 ENST00000372952.7 |
GDAP1L1
|
ganglioside induced differentiation associated protein 1 like 1 |
| chr17_+_11241187 | 1.14 |
ENST00000441885.8
|
SHISA6
|
shisa family member 6 |
| chr12_+_64780465 | 1.14 |
ENST00000542120.6
|
TBC1D30
|
TBC1 domain family member 30 |
| chr12_+_111034136 | 1.11 |
ENST00000261726.11
|
CUX2
|
cut like homeobox 2 |
| chr13_+_79481124 | 1.09 |
ENST00000612570.4
ENST00000218652.11 |
NDFIP2
|
Nedd4 family interacting protein 2 |
| chr6_-_149864300 | 1.09 |
ENST00000239367.7
|
LRP11
|
LDL receptor related protein 11 |
| chr12_-_57633136 | 1.06 |
ENST00000341156.9
ENST00000550764.5 ENST00000551220.1 |
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyltransferase 1 |
| chrX_+_37571569 | 1.05 |
ENST00000614025.4
ENST00000378621.7 ENST00000378619.4 |
LANCL3
|
LanC like 3 |
| chr1_-_154870264 | 1.05 |
ENST00000618040.4
ENST00000271915.9 |
KCNN3
|
potassium calcium-activated channel subfamily N member 3 |
| chrX_+_104166436 | 1.04 |
ENST00000493442.2
|
FAM199X
|
family with sequence similarity 199, X-linked |
| chrX_-_136767322 | 1.04 |
ENST00000370620.5
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr6_+_33420201 | 1.04 |
ENST00000644458.1
ENST00000449372.7 ENST00000628646.2 ENST00000418600.7 |
SYNGAP1
|
synaptic Ras GTPase activating protein 1 |
| chr15_-_45522747 | 1.02 |
ENST00000261867.5
|
SLC30A4
|
solute carrier family 30 member 4 |
| chr12_-_24949026 | 1.00 |
ENST00000539780.5
ENST00000546285.1 ENST00000342945.9 ENST00000261192.12 |
BCAT1
|
branched chain amino acid transaminase 1 |
| chr3_+_142723999 | 1.00 |
ENST00000476941.6
ENST00000273482.10 |
TRPC1
|
transient receptor potential cation channel subfamily C member 1 |
| chr14_-_93788475 | 0.99 |
ENST00000393140.6
|
PRIMA1
|
proline rich membrane anchor 1 |
| chr17_+_74987581 | 0.98 |
ENST00000337231.5
|
CDR2L
|
cerebellar degeneration related protein 2 like |
| chr18_+_34493289 | 0.97 |
ENST00000682923.1
ENST00000596745.5 ENST00000283365.14 ENST00000315456.10 ENST00000598774.6 ENST00000684266.1 ENST00000683092.1 ENST00000683379.1 ENST00000684359.1 |
DTNA
|
dystrobrevin alpha |
| chr13_+_94601830 | 0.97 |
ENST00000376958.5
|
GPR180
|
G protein-coupled receptor 180 |
| chr16_+_53703963 | 0.95 |
ENST00000636218.1
ENST00000637001.1 ENST00000471389.6 ENST00000637969.1 ENST00000640179.1 |
FTO
|
FTO alpha-ketoglutarate dependent dioxygenase |
| chr2_-_212538766 | 0.95 |
ENST00000342788.9
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4 |
| chr4_+_92303946 | 0.94 |
ENST00000282020.9
|
GRID2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr11_-_95924067 | 0.93 |
ENST00000676027.1
ENST00000675489.1 ENST00000409459.5 ENST00000676261.1 ENST00000352297.11 ENST00000346299.10 ENST00000676272.1 ENST00000393223.8 ENST00000675022.1 ENST00000675362.1 ENST00000675174.1 ENST00000674989.1 ENST00000675848.1 ENST00000675652.1 ENST00000481642.6 |
MTMR2
|
myotubularin related protein 2 |
| chr4_+_169620527 | 0.90 |
ENST00000360642.7
ENST00000512813.5 ENST00000513761.6 |
CLCN3
|
chloride voltage-gated channel 3 |
| chr5_+_76083360 | 0.88 |
ENST00000502798.7
|
SV2C
|
synaptic vesicle glycoprotein 2C |
| chr3_-_120094436 | 0.87 |
ENST00000264235.13
ENST00000677034.1 |
GSK3B
|
glycogen synthase kinase 3 beta |
| chrX_-_53281609 | 0.87 |
ENST00000638630.1
ENST00000375365.2 |
IQSEC2
|
IQ motif and Sec7 domain ArfGEF 2 |
| chr18_+_62187247 | 0.87 |
ENST00000644646.2
ENST00000256858.10 ENST00000398130.6 |
RELCH
|
RAB11 binding and LisH domain, coiled-coil and HEAT repeat containing |
| chr1_+_37793865 | 0.86 |
ENST00000397631.7
|
MANEAL
|
mannosidase endo-alpha like |
| chr16_-_80804581 | 0.86 |
ENST00000570137.7
|
CDYL2
|
chromodomain Y like 2 |
| chr8_-_23457618 | 0.86 |
ENST00000358689.9
ENST00000518718.1 |
ENTPD4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr18_+_8717371 | 0.86 |
ENST00000359865.7
|
MTCL1
|
microtubule crosslinking factor 1 |
| chr2_-_121649431 | 0.85 |
ENST00000455322.6
ENST00000409078.8 ENST00000263710.8 ENST00000397587.7 ENST00000541377.5 |
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr21_-_31558977 | 0.85 |
ENST00000286827.7
ENST00000541036.5 |
TIAM1
|
TIAM Rac1 associated GEF 1 |
| chr1_+_96721762 | 0.85 |
ENST00000675735.1
ENST00000609116.5 ENST00000674951.1 ENST00000426398.3 ENST00000370197.5 ENST00000370198.5 |
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr11_+_22193009 | 0.85 |
ENST00000682341.1
ENST00000684663.1 ENST00000683411.1 ENST00000324559.9 |
ANO5
|
anoctamin 5 |
| chrX_+_153687918 | 0.84 |
ENST00000253122.10
|
SLC6A8
|
solute carrier family 6 member 8 |
| chr15_-_34336749 | 0.82 |
ENST00000397707.6
ENST00000560611.5 |
SLC12A6
|
solute carrier family 12 member 6 |
| chr6_+_87155537 | 0.82 |
ENST00000369577.8
ENST00000518845.1 ENST00000339907.8 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr11_-_1572261 | 0.82 |
ENST00000397374.8
|
DUSP8
|
dual specificity phosphatase 8 |
| chr3_+_197960200 | 0.80 |
ENST00000482695.5
ENST00000330198.8 ENST00000419117.5 ENST00000420910.6 ENST00000332636.5 |
LMLN
|
leishmanolysin like peptidase |
| chr7_-_72336995 | 0.79 |
ENST00000329008.9
|
CALN1
|
calneuron 1 |
| chr13_+_31199959 | 0.79 |
ENST00000343307.5
|
B3GLCT
|
beta 3-glucosyltransferase |
| chr2_+_6917404 | 0.78 |
ENST00000320892.11
|
RNF144A
|
ring finger protein 144A |
| chr1_-_230868474 | 0.78 |
ENST00000366663.10
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr6_+_163414637 | 0.77 |
ENST00000453779.6
ENST00000275262.11 ENST00000392127.6 |
QKI
|
QKI, KH domain containing RNA binding |
| chr21_-_39313610 | 0.76 |
ENST00000342449.8
ENST00000341322.4 |
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr1_-_32872473 | 0.75 |
ENST00000496770.1
|
FNDC5
|
fibronectin type III domain containing 5 |
| chr10_-_79445617 | 0.74 |
ENST00000372336.4
|
ZCCHC24
|
zinc finger CCHC-type containing 24 |
| chr1_-_220272415 | 0.74 |
ENST00000358951.7
|
RAB3GAP2
|
RAB3 GTPase activating non-catalytic protein subunit 2 |
| chr10_-_87818153 | 0.71 |
ENST00000308448.11
ENST00000680024.1 |
ATAD1
|
ATPase family AAA domain containing 1 |
| chr2_+_197515565 | 0.70 |
ENST00000233892.8
ENST00000409916.5 |
MOB4
|
MOB family member 4, phocein |
| chr15_-_72117712 | 0.70 |
ENST00000444904.5
ENST00000564571.5 |
MYO9A
|
myosin IXA |
| chr10_-_97292625 | 0.70 |
ENST00000466484.1
ENST00000358531.9 ENST00000358308.7 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr8_-_88327475 | 0.70 |
ENST00000286614.11
|
MMP16
|
matrix metallopeptidase 16 |
| chr15_+_77420880 | 0.69 |
ENST00000336216.9
ENST00000558176.1 |
HMG20A
|
high mobility group 20A |
| chr6_-_93419545 | 0.67 |
ENST00000369297.1
ENST00000369303.9 ENST00000680224.1 ENST00000681532.1 ENST00000679565.1 |
EPHA7
|
EPH receptor A7 |
| chr9_+_15553002 | 0.67 |
ENST00000380701.8
|
CCDC171
|
coiled-coil domain containing 171 |
| chr11_+_99020940 | 0.66 |
ENST00000524871.6
|
CNTN5
|
contactin 5 |
| chr2_-_174634566 | 0.65 |
ENST00000392547.6
|
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr18_+_9475494 | 0.64 |
ENST00000383432.8
|
RALBP1
|
ralA binding protein 1 |
| chr12_+_55973913 | 0.64 |
ENST00000553116.5
|
RAB5B
|
RAB5B, member RAS oncogene family |
| chr17_-_46818680 | 0.63 |
ENST00000225512.6
|
WNT3
|
Wnt family member 3 |
| chr10_+_97426162 | 0.61 |
ENST00000334828.6
|
PGAM1
|
phosphoglycerate mutase 1 |
| chr5_-_36151853 | 0.60 |
ENST00000296603.5
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr12_+_104456962 | 0.60 |
ENST00000547956.1
ENST00000549260.5 |
CHST11
|
carbohydrate sulfotransferase 11 |
| chr15_-_33067884 | 0.60 |
ENST00000334528.13
|
FMN1
|
formin 1 |
| chr7_+_100015588 | 0.60 |
ENST00000324306.11
ENST00000426572.5 |
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr11_-_95231046 | 0.60 |
ENST00000416495.6
ENST00000536441.7 |
SESN3
|
sestrin 3 |
| chr22_+_40951364 | 0.59 |
ENST00000216225.9
|
RBX1
|
ring-box 1 |
| chr19_-_12723925 | 0.59 |
ENST00000425528.6
ENST00000589337.5 ENST00000588216.5 |
TNPO2
|
transportin 2 |
| chr14_+_73644875 | 0.58 |
ENST00000554113.5
ENST00000553645.7 ENST00000555631.6 ENST00000311089.7 ENST00000555919.7 ENST00000554339.5 ENST00000554871.5 |
DNAL1
|
dynein axonemal light chain 1 |
| chr16_-_71724700 | 0.58 |
ENST00000568954.5
|
PHLPP2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr19_+_10420474 | 0.57 |
ENST00000380702.7
|
PDE4A
|
phosphodiesterase 4A |
| chr8_-_142614469 | 0.57 |
ENST00000356613.4
|
ARC
|
activity regulated cytoskeleton associated protein |
| chrX_+_54440396 | 0.57 |
ENST00000375151.5
|
TSR2
|
TSR2 ribosome maturation factor |
| chr5_-_41510554 | 0.56 |
ENST00000377801.8
|
PLCXD3
|
phosphatidylinositol specific phospholipase C X domain containing 3 |
| chr10_+_25174969 | 0.56 |
ENST00000376351.4
|
GPR158
|
G protein-coupled receptor 158 |
| chr1_+_183636065 | 0.56 |
ENST00000304685.8
|
RGL1
|
ral guanine nucleotide dissociation stimulator like 1 |
| chr3_+_169966764 | 0.54 |
ENST00000337002.9
ENST00000480708.5 |
SEC62
|
SEC62 homolog, preprotein translocation factor |
| chr3_-_48685835 | 0.54 |
ENST00000439518.5
ENST00000416649.6 ENST00000294129.7 |
NCKIPSD
|
NCK interacting protein with SH3 domain |
| chr3_-_24494791 | 0.53 |
ENST00000431815.5
ENST00000356447.9 ENST00000418247.1 ENST00000416420.5 ENST00000396671.7 |
THRB
|
thyroid hormone receptor beta |
| chr2_-_106887073 | 0.53 |
ENST00000361686.8
ENST00000409087.3 |
ST6GAL2
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AUGGCUU
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.6 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 1.6 | 4.8 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.9 | 5.7 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.9 | 2.7 | GO:1905033 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.8 | 2.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.6 | 5.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.6 | 1.8 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.6 | 1.8 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.6 | 8.8 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.6 | 15.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.5 | 3.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.5 | 1.5 | GO:1902948 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.5 | 1.4 | GO:0090274 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.3 | 0.3 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.3 | 1.0 | GO:0042245 | RNA repair(GO:0042245) |
| 0.3 | 3.4 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.3 | 4.1 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.3 | 0.8 | GO:1904268 | positive regulation of Schwann cell migration(GO:1900149) regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.3 | 3.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.3 | 0.8 | GO:1902598 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.3 | 8.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 1.6 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.3 | 2.6 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.3 | 1.3 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.2 | 1.7 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.2 | 3.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 0.7 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.2 | 0.9 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.2 | 3.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 | 4.8 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 2.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.2 | 1.9 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 1.7 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 1.7 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.2 | 4.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 1.6 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.2 | 3.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.2 | 2.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 1.0 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 4.9 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.2 | 2.5 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 2.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 1.0 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.2 | 1.7 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 5.8 | GO:0060384 | innervation(GO:0060384) |
| 0.2 | 3.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 0.9 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.2 | 3.0 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.2 | 1.9 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.2 | 1.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 0.5 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.2 | 0.6 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.2 | 0.3 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.2 | 1.6 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.2 | 7.5 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.2 | 2.1 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.6 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.1 | 2.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 6.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 6.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.9 | GO:2000077 | negative regulation of dopaminergic neuron differentiation(GO:1904339) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.1 | 0.9 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.1 | 2.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.5 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.1 | 1.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.8 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.5 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.1 | 0.6 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 | 1.7 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 0.6 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.3 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 1.7 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.1 | 7.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 2.3 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 6.1 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 0.8 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 1.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.1 | 0.6 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.9 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 0.5 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.5 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.9 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) |
| 0.1 | 0.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 1.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.4 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 1.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 2.0 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.3 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.0 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 0.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.8 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 1.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 3.8 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 1.6 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 2.5 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.4 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 4.4 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.5 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.3 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 5.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.9 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 1.1 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0060382 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.7 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 1.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.7 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis(GO:0060978) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.2 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 1.0 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 2.0 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 1.7 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.8 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 1.9 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 1.2 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 1.2 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.3 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.0 | 0.7 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.3 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 1.0 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 1.0 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.4 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.0 | 1.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.5 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.0 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.0 | 0.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 1.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.3 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.0 | 0.5 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.0 | GO:1900238 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.3 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.5 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.2 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.6 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 2.1 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 3.8 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.4 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 4.3 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 0.1 | GO:0072423 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 1.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 3.0 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.8 | GO:0098855 | HCN channel complex(GO:0098855) |
| 1.7 | 13.3 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 1.6 | 4.8 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 0.8 | 2.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.7 | 2.0 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.6 | 5.7 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.6 | 5.9 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.5 | 1.6 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.5 | 2.7 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.5 | 8.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.4 | 3.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.3 | 13.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 8.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 0.9 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 3.4 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.2 | 1.7 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.2 | 0.9 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.2 | 5.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 8.9 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 1.7 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 2.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 5.0 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 4.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.6 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 1.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 5.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 5.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 5.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 2.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 1.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 2.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 6.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.9 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 1.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 4.4 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 1.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 5.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.9 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 4.4 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 7.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.2 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.7 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 5.7 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 5.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.5 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 1.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 6.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 3.7 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.0 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 1.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 8.8 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.1 | 7.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 1.0 | 4.8 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.8 | 2.3 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.6 | 3.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.6 | 5.7 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.5 | 5.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.5 | 2.0 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.5 | 1.9 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.4 | 4.5 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.4 | 2.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.4 | 20.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.4 | 1.6 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.4 | 2.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.4 | 3.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.3 | 5.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 9.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.3 | 0.8 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.3 | 1.1 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.2 | 1.7 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.2 | 1.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.2 | 3.0 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 0.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 1.0 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 1.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 1.7 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.2 | 1.7 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.2 | 2.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 4.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 1.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 2.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 3.9 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 4.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 3.5 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 4.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.4 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 3.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 4.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 6.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.6 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 1.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.9 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 4.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 1.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 1.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.6 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 1.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 4.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.4 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 2.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 2.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.2 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 0.6 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 1.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.3 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 1.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 2.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 0.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 1.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 5.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.8 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 1.0 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 1.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.7 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 9.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 1.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.0 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 1.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.9 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 0.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 2.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0052848 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 8.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 5.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 3.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 3.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.2 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 0.2 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 2.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.0 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 7.9 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 5.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 7.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 2.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 2.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 3.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 3.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 13.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 0.6 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.2 | 7.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 12.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 12.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 5.0 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 2.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 17.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 3.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 2.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 10.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 2.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 2.3 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.2 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |