Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
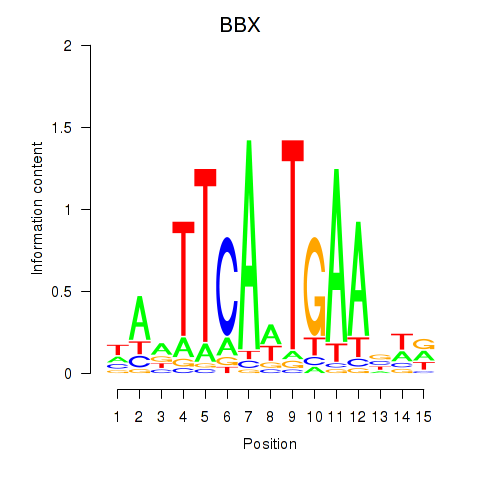
Results for BBX
Z-value: 0.89
Transcription factors associated with BBX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BBX
|
ENSG00000114439.19 | BBX high mobility group box domain containing |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BBX | hg38_v1_chr3_+_107524357_107524376 | 0.11 | 5.5e-01 | Click! |
Activity profile of BBX motif
Sorted Z-values of BBX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_154563003 | 5.60 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr7_-_16881967 | 3.47 |
ENST00000402239.7
ENST00000310398.7 ENST00000414935.1 |
AGR3
|
anterior gradient 3, protein disulphide isomerase family member |
| chr16_+_56638659 | 3.18 |
ENST00000290705.12
|
MT1A
|
metallothionein 1A |
| chr11_+_60699605 | 3.18 |
ENST00000300226.7
|
MS4A8
|
membrane spanning 4-domains A8 |
| chr2_-_215393126 | 2.53 |
ENST00000456923.5
|
FN1
|
fibronectin 1 |
| chr9_-_92404559 | 2.49 |
ENST00000262551.8
ENST00000375561.10 |
OGN
|
osteoglycin |
| chr7_-_16833411 | 2.41 |
ENST00000412973.1
|
AGR2
|
anterior gradient 2, protein disulphide isomerase family member |
| chr11_+_60699669 | 2.27 |
ENST00000529752.5
|
MS4A8
|
membrane spanning 4-domains A8 |
| chr3_-_58666765 | 2.27 |
ENST00000358781.7
|
FAM3D
|
FAM3 metabolism regulating signaling molecule D |
| chr2_+_102311546 | 2.20 |
ENST00000233954.6
ENST00000447231.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr9_-_92404690 | 2.06 |
ENST00000447356.1
|
OGN
|
osteoglycin |
| chr3_+_151814102 | 1.98 |
ENST00000232892.12
|
AADAC
|
arylacetamide deacetylase |
| chr1_-_58577244 | 1.91 |
ENST00000371225.4
|
TACSTD2
|
tumor associated calcium signal transducer 2 |
| chr7_+_117480052 | 1.65 |
ENST00000426809.5
ENST00000649781.1 |
CFTR
|
CF transmembrane conductance regulator |
| chr2_+_102311502 | 1.63 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr6_-_25874212 | 1.56 |
ENST00000361703.10
ENST00000397060.8 |
SLC17A3
|
solute carrier family 17 member 3 |
| chr7_+_117480011 | 1.52 |
ENST00000649406.1
ENST00000648260.1 ENST00000003084.11 |
CFTR
|
CF transmembrane conductance regulator |
| chr9_+_35161951 | 1.51 |
ENST00000617908.4
ENST00000619578.4 |
UNC13B
|
unc-13 homolog B |
| chr3_+_148865288 | 1.35 |
ENST00000296046.4
|
CPA3
|
carboxypeptidase A3 |
| chr6_+_31927486 | 1.33 |
ENST00000442278.6
|
C2
|
complement C2 |
| chr3_-_169147734 | 1.30 |
ENST00000464456.5
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr4_-_185775271 | 1.29 |
ENST00000430503.5
ENST00000319454.10 ENST00000450341.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_-_49744434 | 1.25 |
ENST00000433368.6
ENST00000354620.4 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr6_+_121437378 | 1.19 |
ENST00000650427.1
ENST00000647564.1 |
GJA1
|
gap junction protein alpha 1 |
| chr6_-_49744378 | 1.18 |
ENST00000371159.8
ENST00000263045.9 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr4_-_103099811 | 1.14 |
ENST00000504285.5
ENST00000296424.9 |
BDH2
|
3-hydroxybutyrate dehydrogenase 2 |
| chr3_+_151814069 | 1.13 |
ENST00000488869.1
|
AADAC
|
arylacetamide deacetylase |
| chr4_-_185775890 | 1.11 |
ENST00000437304.6
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_-_146528750 | 1.07 |
ENST00000483300.5
|
PLSCR1
|
phospholipid scramblase 1 |
| chr9_+_35162000 | 1.06 |
ENST00000396787.5
ENST00000378495.7 ENST00000635942.1 ENST00000378496.8 |
UNC13B
|
unc-13 homolog B |
| chr15_+_49423233 | 1.05 |
ENST00000560270.1
ENST00000267843.9 ENST00000560979.1 |
FGF7
|
fibroblast growth factor 7 |
| chr15_+_40351026 | 0.99 |
ENST00000448599.2
|
PHGR1
|
proline, histidine and glycine rich 1 |
| chr4_-_103099852 | 0.99 |
ENST00000509245.1
|
BDH2
|
3-hydroxybutyrate dehydrogenase 2 |
| chr9_-_92482499 | 0.99 |
ENST00000375544.7
|
ASPN
|
asporin |
| chr6_+_31927510 | 0.97 |
ENST00000447952.6
|
C2
|
complement C2 |
| chr1_+_192575765 | 0.95 |
ENST00000469578.2
ENST00000367459.8 |
RGS1
|
regulator of G protein signaling 1 |
| chr4_-_185812209 | 0.95 |
ENST00000393523.6
ENST00000393528.7 ENST00000449407.6 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr19_+_48606732 | 0.94 |
ENST00000321762.3
|
SPACA4
|
sperm acrosome associated 4 |
| chr20_-_7940444 | 0.92 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1 |
| chr20_-_46684467 | 0.92 |
ENST00000372121.5
|
SLC13A3
|
solute carrier family 13 member 3 |
| chr5_+_141135199 | 0.89 |
ENST00000231134.8
ENST00000623915.1 |
PCDHB5
|
protocadherin beta 5 |
| chr4_-_185775484 | 0.88 |
ENST00000444771.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_+_66075876 | 0.87 |
ENST00000395332.8
|
ASL
|
argininosuccinate lyase |
| chr9_-_92482350 | 0.87 |
ENST00000375543.2
|
ASPN
|
asporin |
| chr4_-_185775432 | 0.84 |
ENST00000457247.5
ENST00000435480.5 ENST00000425679.5 ENST00000457934.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_36084079 | 0.84 |
ENST00000207457.8
|
TEKT2
|
tektin 2 |
| chr7_+_32979445 | 0.83 |
ENST00000418354.5
ENST00000490776.3 |
FKBP9
|
FKBP prolyl isomerase 9 |
| chr4_-_185775411 | 0.79 |
ENST00000445115.5
ENST00000451701.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr8_-_28490220 | 0.78 |
ENST00000517673.5
ENST00000380254.7 ENST00000518734.5 ENST00000346498.6 |
FBXO16
|
F-box protein 16 |
| chr4_-_185775376 | 0.62 |
ENST00000456596.5
ENST00000414724.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_+_52420107 | 0.60 |
ENST00000636489.1
ENST00000637089.1 ENST00000637353.1 ENST00000637263.1 |
EFHC1
|
EF-hand domain containing 1 |
| chr12_+_18262730 | 0.60 |
ENST00000675017.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr6_+_151036912 | 0.57 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr12_-_39340963 | 0.55 |
ENST00000552961.5
|
KIF21A
|
kinesin family member 21A |
| chr4_+_107824555 | 0.54 |
ENST00000394684.8
|
SGMS2
|
sphingomyelin synthase 2 |
| chr9_-_2844058 | 0.53 |
ENST00000397885.3
|
PUM3
|
pumilio RNA binding family member 3 |
| chr5_+_141430565 | 0.52 |
ENST00000613314.1
|
PCDHGA12
|
protocadherin gamma subfamily A, 12 |
| chr3_+_130440510 | 0.51 |
ENST00000373157.8
|
COL6A5
|
collagen type VI alpha 5 chain |
| chr8_-_144605699 | 0.51 |
ENST00000377307.6
ENST00000276826.5 |
ARHGAP39
|
Rho GTPase activating protein 39 |
| chr2_+_200440649 | 0.49 |
ENST00000366118.2
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr2_+_100821025 | 0.47 |
ENST00000427413.5
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr8_-_6926066 | 0.47 |
ENST00000297436.3
|
DEFA6
|
defensin alpha 6 |
| chr17_+_42798881 | 0.45 |
ENST00000588527.5
|
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr1_+_54998927 | 0.44 |
ENST00000651561.1
|
BSND
|
barttin CLCNK type accessory subunit beta |
| chr6_-_87291988 | 0.41 |
ENST00000369576.2
|
GJB7
|
gap junction protein beta 7 |
| chr5_+_141430499 | 0.40 |
ENST00000252085.4
|
PCDHGA12
|
protocadherin gamma subfamily A, 12 |
| chr3_-_185821092 | 0.39 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr5_+_102864505 | 0.37 |
ENST00000511839.5
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr3_+_122680802 | 0.35 |
ENST00000474629.7
|
PARP14
|
poly(ADP-ribose) polymerase family member 14 |
| chr1_-_161367872 | 0.35 |
ENST00000367974.2
|
CFAP126
|
cilia and flagella associated protein 126 |
| chrX_-_126166273 | 0.34 |
ENST00000360028.4
|
DCAF12L2
|
DDB1 and CUL4 associated factor 12 like 2 |
| chr6_+_54308429 | 0.32 |
ENST00000259782.9
ENST00000370864.3 |
TINAG
|
tubulointerstitial nephritis antigen |
| chr4_+_87650277 | 0.32 |
ENST00000339673.11
ENST00000282479.8 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr4_-_185761562 | 0.31 |
ENST00000445343.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_+_128709052 | 0.31 |
ENST00000485070.5
|
FAM71F1
|
family with sequence similarity 71 member F1 |
| chr9_-_13175824 | 0.29 |
ENST00000545857.5
|
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr16_-_58734299 | 0.28 |
ENST00000245206.10
ENST00000434819.2 |
GOT2
|
glutamic-oxaloacetic transaminase 2 |
| chr6_+_117482634 | 0.28 |
ENST00000296955.12
ENST00000338728.10 |
DCBLD1
|
discoidin, CUB and LCCL domain containing 1 |
| chr9_+_101185029 | 0.27 |
ENST00000395056.2
|
PLPPR1
|
phospholipid phosphatase related 1 |
| chr15_+_72118392 | 0.26 |
ENST00000340912.6
|
SENP8
|
SUMO peptidase family member, NEDD8 specific |
| chr5_+_141182369 | 0.25 |
ENST00000609684.3
ENST00000625044.1 ENST00000623407.1 ENST00000623884.1 |
PCDHB16
ENSG00000279068.1
|
protocadherin beta 16 novel transcript |
| chr5_+_126423122 | 0.24 |
ENST00000515200.5
|
GRAMD2B
|
GRAM domain containing 2B |
| chr20_-_31722854 | 0.24 |
ENST00000307677.5
|
BCL2L1
|
BCL2 like 1 |
| chr14_+_67189393 | 0.23 |
ENST00000524532.5
ENST00000612183.4 ENST00000530728.5 |
FAM71D
|
family with sequence similarity 71 member D |
| chr16_+_56191476 | 0.23 |
ENST00000262493.12
|
GNAO1
|
G protein subunit alpha o1 |
| chr9_-_5833014 | 0.22 |
ENST00000339450.10
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr14_+_20110739 | 0.20 |
ENST00000641386.2
ENST00000641633.2 |
OR4K17
|
olfactory receptor family 4 subfamily K member 17 |
| chr4_+_68447453 | 0.20 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr6_+_24356903 | 0.19 |
ENST00000274766.2
|
KAAG1
|
kidney associated antigen 1 |
| chr6_-_135497230 | 0.19 |
ENST00000681365.1
|
AHI1
|
Abelson helper integration site 1 |
| chr8_-_92019156 | 0.19 |
ENST00000521054.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr14_+_37795308 | 0.19 |
ENST00000267368.11
ENST00000382320.4 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr12_-_100262356 | 0.19 |
ENST00000548313.5
|
DEPDC4
|
DEP domain containing 4 |
| chr20_-_31722949 | 0.18 |
ENST00000376055.9
|
BCL2L1
|
BCL2 like 1 |
| chr11_-_14520429 | 0.18 |
ENST00000533068.5
|
PSMA1
|
proteasome 20S subunit alpha 1 |
| chr2_-_61854013 | 0.17 |
ENST00000404929.6
|
FAM161A
|
FAM161 centrosomal protein A |
| chr11_-_63229652 | 0.17 |
ENST00000306494.10
|
SLC22A25
|
solute carrier family 22 member 25 |
| chrX_-_141698739 | 0.17 |
ENST00000370515.3
|
SPANXD
|
SPANX family member D |
| chrX_+_54807599 | 0.17 |
ENST00000375053.6
ENST00000627068.2 ENST00000375068.6 ENST00000347546.8 |
MAGED2
|
MAGE family member D2 |
| chr2_-_61854119 | 0.17 |
ENST00000405894.3
|
FAM161A
|
FAM161 centrosomal protein A |
| chr4_-_993430 | 0.16 |
ENST00000361661.6
ENST00000622731.4 |
SLC26A1
|
solute carrier family 26 member 1 |
| chr9_-_104599413 | 0.16 |
ENST00000374779.3
|
OR13C5
|
olfactory receptor family 13 subfamily C member 5 |
| chr4_-_185813121 | 0.15 |
ENST00000456060.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr21_+_30396030 | 0.15 |
ENST00000355459.4
|
KRTAP13-1
|
keratin associated protein 13-1 |
| chr6_+_132538290 | 0.14 |
ENST00000434551.2
|
TAAR9
|
trace amine associated receptor 9 |
| chr6_-_160664270 | 0.14 |
ENST00000316300.10
|
LPA
|
lipoprotein(a) |
| chr1_+_51102221 | 0.13 |
ENST00000648827.1
ENST00000371759.7 |
C1orf185
|
chromosome 1 open reading frame 185 |
| chr8_-_107497909 | 0.13 |
ENST00000517746.6
|
ANGPT1
|
angiopoietin 1 |
| chr8_-_107498041 | 0.12 |
ENST00000297450.7
|
ANGPT1
|
angiopoietin 1 |
| chr4_+_94489030 | 0.11 |
ENST00000510099.5
|
PDLIM5
|
PDZ and LIM domain 5 |
| chr18_-_13915531 | 0.10 |
ENST00000327606.4
|
MC2R
|
melanocortin 2 receptor |
| chr1_+_51102238 | 0.10 |
ENST00000467127.5
|
C1orf185
|
chromosome 1 open reading frame 185 |
| chr9_+_65700287 | 0.10 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr2_-_119366682 | 0.10 |
ENST00000409877.5
ENST00000409523.1 ENST00000409466.6 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr19_+_7763210 | 0.10 |
ENST00000359059.10
ENST00000596363.5 ENST00000394122.7 ENST00000327325.10 |
CLEC4M
|
C-type lectin domain family 4 member M |
| chr2_+_196713117 | 0.09 |
ENST00000409270.5
|
CCDC150
|
coiled-coil domain containing 150 |
| chr1_-_153150884 | 0.09 |
ENST00000368748.5
|
SPRR2G
|
small proline rich protein 2G |
| chr4_+_69280472 | 0.09 |
ENST00000335568.10
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase family 2 member B28 |
| chr4_+_158315309 | 0.09 |
ENST00000460056.6
|
RXFP1
|
relaxin family peptide receptor 1 |
| chr13_+_27919993 | 0.08 |
ENST00000381033.5
|
PDX1
|
pancreatic and duodenal homeobox 1 |
| chr1_-_686673 | 0.07 |
ENST00000332831.4
|
OR4F16
|
olfactory receptor family 4 subfamily F member 16 |
| chr1_+_170145959 | 0.07 |
ENST00000439373.3
|
METTL11B
|
methyltransferase like 11B |
| chr3_+_51817596 | 0.07 |
ENST00000456080.5
|
ENSG00000285749.2
|
novel protein |
| chr19_-_14848922 | 0.07 |
ENST00000641129.1
|
OR7A10
|
olfactory receptor family 7 subfamily A member 10 |
| chr14_+_94026314 | 0.07 |
ENST00000203664.10
ENST00000553723.1 |
OTUB2
|
OTU deubiquitinase, ubiquitin aldehyde binding 2 |
| chr1_+_51617079 | 0.06 |
ENST00000447887.5
ENST00000428468.6 ENST00000453295.5 |
OSBPL9
|
oxysterol binding protein like 9 |
| chr12_-_16600703 | 0.06 |
ENST00000616247.4
|
LMO3
|
LIM domain only 3 |
| chr6_+_29457155 | 0.06 |
ENST00000377133.6
ENST00000377136.5 |
OR2H1
|
olfactory receptor family 2 subfamily H member 1 |
| chr3_+_130850585 | 0.06 |
ENST00000505330.5
ENST00000504381.5 ENST00000507488.6 |
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr4_+_70592253 | 0.06 |
ENST00000322937.10
ENST00000613447.4 |
AMBN
|
ameloblastin |
| chr4_-_993376 | 0.05 |
ENST00000398520.6
ENST00000398516.3 |
SLC26A1
|
solute carrier family 26 member 1 |
| chr4_+_113145608 | 0.05 |
ENST00000511380.1
|
ANK2
|
ankyrin 2 |
| chr18_-_23437927 | 0.05 |
ENST00000578520.5
ENST00000383233.8 |
TMEM241
|
transmembrane protein 241 |
| chr3_+_128051610 | 0.04 |
ENST00000464451.5
|
SEC61A1
|
SEC61 translocon subunit alpha 1 |
| chr20_-_31722533 | 0.04 |
ENST00000677194.1
ENST00000434194.2 ENST00000376062.6 |
BCL2L1
|
BCL2 like 1 |
| chr10_-_84225537 | 0.03 |
ENST00000372113.7
ENST00000538192.5 |
LRIT2
|
leucine rich repeat, Ig-like and transmembrane domains 2 |
| chr3_+_141262614 | 0.03 |
ENST00000504264.5
|
PXYLP1
|
2-phosphoxylose phosphatase 1 |
| chrX_+_11293411 | 0.02 |
ENST00000348912.4
ENST00000380714.7 ENST00000380712.7 |
AMELX
|
amelogenin X-linked |
| chr4_+_26576670 | 0.02 |
ENST00000512840.5
|
TBC1D19
|
TBC1 domain family member 19 |
| chr1_-_13285154 | 0.01 |
ENST00000357367.6
ENST00000614831.1 |
PRAMEF8
|
PRAME family member 8 |
| chr14_+_63761836 | 0.01 |
ENST00000674003.1
|
SYNE2
|
spectrin repeat containing nuclear envelope protein 2 |
| chr15_-_33119365 | 0.01 |
ENST00000672206.1
|
FMN1
|
formin 1 |
| chr1_+_95151377 | 0.01 |
ENST00000604203.1
|
TLCD4-RWDD3
|
TLCD4-RWDD3 readthrough |
| chr5_+_171785817 | 0.01 |
ENST00000523047.4
ENST00000330910.7 |
SMIM23
|
small integral membrane protein 23 |
| chr15_-_72118114 | 0.00 |
ENST00000356056.10
ENST00000569314.1 |
MYO9A
|
myosin IXA |
Network of associatons between targets according to the STRING database.
First level regulatory network of BBX
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.8 | 2.5 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.8 | 3.2 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.5 | 2.4 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.5 | 3.8 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.4 | 5.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.4 | 1.1 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.3 | 0.9 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.3 | 1.2 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.2 | 1.9 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.2 | 0.9 | GO:0015744 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.2 | 4.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.2 | 3.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 | 0.9 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.2 | 1.9 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.2 | 1.6 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 2.3 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 2.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 7.1 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.6 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.9 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 3.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.3 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.1 | 0.3 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.1 | 0.3 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.5 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.9 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.4 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 1.4 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.0 | 0.2 | GO:0039007 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.5 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.8 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 2.3 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 1.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 1.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.5 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.2 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.1 | GO:0010260 | organ senescence(GO:0010260) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.0 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.6 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.5 | 3.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.3 | 2.6 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 1.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 5.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.4 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 5.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 6.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.8 | 3.2 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.5 | 2.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.3 | 0.9 | GO:0003973 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.3 | 0.9 | GO:0004056 | argininosuccinate lyase activity(GO:0004056) |
| 0.2 | 0.9 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.2 | 1.6 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 5.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 6.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 0.6 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.3 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 3.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 2.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.4 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.9 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 8.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 1.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 1.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 4.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.9 | GO:0005518 | collagen binding(GO:0005518) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 8.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 4.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 2.5 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 2.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.1 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |