Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
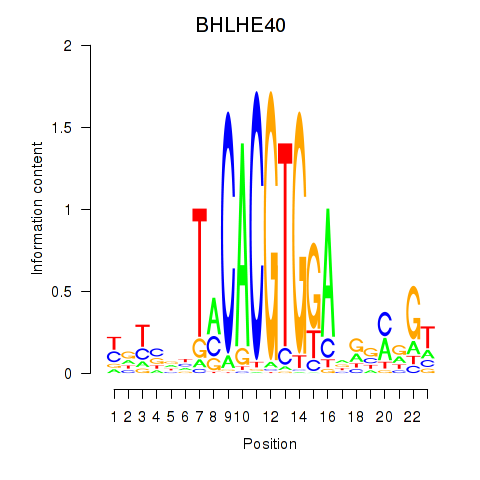
Results for BHLHE40
Z-value: 0.72
Transcription factors associated with BHLHE40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BHLHE40
|
ENSG00000134107.5 | basic helix-loop-helix family member e40 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BHLHE40 | hg38_v1_chr3_+_4979428_4979495 | -0.22 | 2.2e-01 | Click! |
Activity profile of BHLHE40 motif
Sorted Z-values of BHLHE40 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_92923143 | 2.72 |
ENST00000216492.10
ENST00000334654.4 |
CHGA
|
chromogranin A |
| chr6_-_136550819 | 1.42 |
ENST00000616617.4
ENST00000618822.4 |
MAP7
|
microtubule associated protein 7 |
| chr8_-_98117155 | 1.19 |
ENST00000254878.8
ENST00000521560.1 |
RIDA
|
reactive intermediate imine deaminase A homolog |
| chr22_+_48489546 | 1.14 |
ENST00000402357.6
|
TAFA5
|
TAFA chemokine like family member 5 |
| chr8_-_98117110 | 1.05 |
ENST00000520507.5
|
RIDA
|
reactive intermediate imine deaminase A homolog |
| chr17_-_63773534 | 1.01 |
ENST00000403162.7
ENST00000582252.1 ENST00000225726.10 |
CCDC47
|
coiled-coil domain containing 47 |
| chr2_-_228181669 | 1.00 |
ENST00000392056.8
|
SPHKAP
|
SPHK1 interactor, AKAP domain containing |
| chr11_+_69155478 | 1.00 |
ENST00000641568.1
|
SMIM38
|
small integral membrane protein 38 |
| chr2_-_228181612 | 0.94 |
ENST00000344657.5
|
SPHKAP
|
SPHK1 interactor, AKAP domain containing |
| chr7_+_148133684 | 0.90 |
ENST00000628930.2
|
CNTNAP2
|
contactin associated protein 2 |
| chr9_+_706841 | 0.88 |
ENST00000382293.7
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chrX_-_2968236 | 0.88 |
ENST00000684117.1
ENST00000672761.1 ENST00000672027.1 ENST00000672606.1 ENST00000673032.1 ENST00000540563.6 |
ARSL
|
arylsulfatase L |
| chr8_-_144605699 | 0.87 |
ENST00000377307.6
ENST00000276826.5 |
ARHGAP39
|
Rho GTPase activating protein 39 |
| chr15_+_68631988 | 0.87 |
ENST00000543950.3
|
CORO2B
|
coronin 2B |
| chr6_-_136550407 | 0.83 |
ENST00000354570.8
|
MAP7
|
microtubule associated protein 7 |
| chr17_+_42659264 | 0.81 |
ENST00000251412.8
|
TUBG2
|
tubulin gamma 2 |
| chr9_+_129140004 | 0.79 |
ENST00000436883.5
ENST00000414510.5 |
PTPA
|
protein phosphatase 2 phosphatase activator |
| chr1_+_43172324 | 0.78 |
ENST00000528956.5
ENST00000610710.4 ENST00000372492.9 ENST00000529956.5 |
CFAP57
|
cilia and flagella associated protein 57 |
| chr17_+_63773863 | 0.77 |
ENST00000578681.5
ENST00000583590.5 |
DDX42
|
DEAD-box helicase 42 |
| chr1_-_111503622 | 0.76 |
ENST00000369716.9
ENST00000241356.5 |
TMIGD3
ADORA3
|
transmembrane and immunoglobulin domain containing 3 adenosine A3 receptor |
| chr2_-_135985866 | 0.76 |
ENST00000441323.5
ENST00000449218.5 |
DARS1
|
aspartyl-tRNA synthetase 1 |
| chr16_+_29900474 | 0.73 |
ENST00000308748.10
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr1_-_43172504 | 0.71 |
ENST00000431635.6
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr11_-_3218813 | 0.70 |
ENST00000332314.3
|
MRGPRG
|
MAS related GPR family member G |
| chr16_+_29900345 | 0.68 |
ENST00000563177.5
ENST00000483405.5 |
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr19_-_23003134 | 0.68 |
ENST00000594710.2
|
ZNF728
|
zinc finger protein 728 |
| chr13_+_100089015 | 0.67 |
ENST00000376286.8
ENST00000376279.7 ENST00000376285.6 |
PCCA
|
propionyl-CoA carboxylase subunit alpha |
| chr1_+_43979877 | 0.67 |
ENST00000356836.10
ENST00000309519.8 |
B4GALT2
|
beta-1,4-galactosyltransferase 2 |
| chr1_-_43172244 | 0.66 |
ENST00000236051.3
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr3_-_179071742 | 0.66 |
ENST00000311417.7
ENST00000652290.1 |
ZMAT3
|
zinc finger matrin-type 3 |
| chr18_+_58045642 | 0.65 |
ENST00000676223.1
ENST00000675147.1 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr21_-_25735026 | 0.64 |
ENST00000400099.5
ENST00000457143.6 |
ATP5PF
|
ATP synthase peripheral stalk subunit F6 |
| chr5_+_121851876 | 0.63 |
ENST00000321339.3
|
FTMT
|
ferritin mitochondrial |
| chr13_+_34942263 | 0.61 |
ENST00000379939.7
ENST00000400445.7 |
NBEA
|
neurobeachin |
| chr5_-_36151853 | 0.61 |
ENST00000296603.5
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr17_-_17206264 | 0.61 |
ENST00000321560.4
|
PLD6
|
phospholipase D family member 6 |
| chr21_-_25735570 | 0.61 |
ENST00000400090.7
ENST00000400087.7 ENST00000400093.3 |
ATP5PF
|
ATP synthase peripheral stalk subunit F6 |
| chr21_-_29019314 | 0.60 |
ENST00000493196.2
|
RWDD2B
|
RWD domain containing 2B |
| chr22_-_30557586 | 0.60 |
ENST00000338911.6
ENST00000453479.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr17_+_63774144 | 0.60 |
ENST00000389924.7
ENST00000359353.9 |
DDX42
|
DEAD-box helicase 42 |
| chr3_+_99817849 | 0.59 |
ENST00000421999.8
|
CMSS1
|
cms1 ribosomal small subunit homolog |
| chr1_+_117929720 | 0.59 |
ENST00000369441.7
ENST00000349139.6 |
WDR3
|
WD repeat domain 3 |
| chr19_+_16661121 | 0.56 |
ENST00000187762.7
ENST00000599479.1 |
TMEM38A
|
transmembrane protein 38A |
| chr8_+_11284789 | 0.55 |
ENST00000221086.8
|
MTMR9
|
myotubularin related protein 9 |
| chr14_+_77458032 | 0.54 |
ENST00000535854.6
ENST00000555517.1 ENST00000216479.8 |
AHSA1
|
activator of HSP90 ATPase activity 1 |
| chr8_-_71547626 | 0.54 |
ENST00000647540.1
ENST00000644229.1 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr6_+_151239951 | 0.53 |
ENST00000402676.7
|
AKAP12
|
A-kinase anchoring protein 12 |
| chr22_+_48489567 | 0.52 |
ENST00000336769.9
|
TAFA5
|
TAFA chemokine like family member 5 |
| chr12_-_24949026 | 0.52 |
ENST00000539780.5
ENST00000546285.1 ENST00000342945.9 ENST00000261192.12 |
BCAT1
|
branched chain amino acid transaminase 1 |
| chr20_-_44311142 | 0.51 |
ENST00000396825.4
|
FITM2
|
fat storage inducing transmembrane protein 2 |
| chr6_-_36874783 | 0.51 |
ENST00000373699.6
|
PPIL1
|
peptidylprolyl isomerase like 1 |
| chr11_+_73308237 | 0.48 |
ENST00000263674.4
|
ARHGEF17
|
Rho guanine nucleotide exchange factor 17 |
| chr1_+_119414931 | 0.48 |
ENST00000543831.5
ENST00000433745.5 ENST00000369416.4 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr8_+_54135203 | 0.48 |
ENST00000260102.9
ENST00000519831.5 |
MRPL15
|
mitochondrial ribosomal protein L15 |
| chr18_+_580367 | 0.48 |
ENST00000327228.5
|
CETN1
|
centrin 1 |
| chr2_+_105038036 | 0.47 |
ENST00000258455.8
|
MRPS9
|
mitochondrial ribosomal protein S9 |
| chr11_+_60842095 | 0.46 |
ENST00000227520.10
|
CCDC86
|
coiled-coil domain containing 86 |
| chr1_+_75796867 | 0.46 |
ENST00000263187.4
|
MSH4
|
mutS homolog 4 |
| chr2_+_102619531 | 0.46 |
ENST00000233969.3
|
SLC9A2
|
solute carrier family 9 member A2 |
| chr3_+_131381671 | 0.45 |
ENST00000537561.5
ENST00000521288.2 ENST00000502852.1 |
NUDT16
|
nudix hydrolase 16 |
| chr2_-_178478499 | 0.44 |
ENST00000434643.6
|
FKBP7
|
FKBP prolyl isomerase 7 |
| chr19_-_5719849 | 0.44 |
ENST00000590729.5
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr6_+_122399621 | 0.44 |
ENST00000368455.9
|
HSF2
|
heat shock transcription factor 2 |
| chr19_-_36573243 | 0.43 |
ENST00000334116.7
ENST00000591340.6 |
ZNF529
|
zinc finger protein 529 |
| chr8_+_108443601 | 0.43 |
ENST00000524143.5
ENST00000220853.8 |
EMC2
|
ER membrane protein complex subunit 2 |
| chr3_+_99817818 | 0.43 |
ENST00000463526.1
|
CMSS1
|
cms1 ribosomal small subunit homolog |
| chrX_-_16869840 | 0.42 |
ENST00000380084.8
|
RBBP7
|
RB binding protein 7, chromatin remodeling factor |
| chr19_-_5720159 | 0.42 |
ENST00000593119.5
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr17_+_21376321 | 0.41 |
ENST00000583088.6
|
KCNJ12
|
potassium inwardly rectifying channel subfamily J member 12 |
| chr22_-_50085331 | 0.40 |
ENST00000395876.6
|
MLC1
|
modulator of VRAC current 1 |
| chr19_-_5720131 | 0.39 |
ENST00000587365.1
ENST00000360614.8 ENST00000585374.5 |
LONP1
|
lon peptidase 1, mitochondrial |
| chr1_-_193105373 | 0.39 |
ENST00000367439.8
|
GLRX2
|
glutaredoxin 2 |
| chr6_+_122399536 | 0.38 |
ENST00000452194.5
|
HSF2
|
heat shock transcription factor 2 |
| chr7_+_116672357 | 0.38 |
ENST00000456159.1
|
MET
|
MET proto-oncogene, receptor tyrosine kinase |
| chr7_+_116672187 | 0.38 |
ENST00000318493.11
ENST00000397752.8 |
MET
|
MET proto-oncogene, receptor tyrosine kinase |
| chr19_-_55140922 | 0.38 |
ENST00000589745.5
|
TNNT1
|
troponin T1, slow skeletal type |
| chr1_-_46941425 | 0.38 |
ENST00000371904.8
|
CYP4A11
|
cytochrome P450 family 4 subfamily A member 11 |
| chr16_-_3099228 | 0.37 |
ENST00000572431.1
ENST00000572548.1 ENST00000575108.5 ENST00000576985.6 ENST00000576483.1 ENST00000538082.5 |
ZSCAN10
|
zinc finger and SCAN domain containing 10 |
| chr21_-_43659460 | 0.37 |
ENST00000443485.1
ENST00000291560.7 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr4_+_56436131 | 0.36 |
ENST00000399688.7
|
PAICS
|
phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase |
| chr17_-_7290392 | 0.36 |
ENST00000571464.1
|
YBX2
|
Y-box binding protein 2 |
| chr5_+_90474848 | 0.35 |
ENST00000651687.1
|
POLR3G
|
RNA polymerase III subunit G |
| chr17_+_2796404 | 0.35 |
ENST00000366401.8
ENST00000254695.13 ENST00000542807.1 |
RAP1GAP2
|
RAP1 GTPase activating protein 2 |
| chr18_+_58045683 | 0.34 |
ENST00000592846.5
ENST00000675801.1 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr17_+_74432079 | 0.34 |
ENST00000392627.7
ENST00000392628.7 ENST00000582473.2 |
GPRC5C
|
G protein-coupled receptor class C group 5 member C |
| chr6_-_111606260 | 0.33 |
ENST00000340026.10
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr5_-_145835285 | 0.33 |
ENST00000505416.5
ENST00000334744.8 ENST00000511435.1 |
PRELID2
|
PRELI domain containing 2 |
| chr2_-_178478541 | 0.33 |
ENST00000424785.7
|
FKBP7
|
FKBP prolyl isomerase 7 |
| chr10_-_48251757 | 0.33 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr17_-_54968637 | 0.32 |
ENST00000299335.8
|
COX11
|
cytochrome c oxidase copper chaperone COX11 |
| chr8_-_30912998 | 0.32 |
ENST00000643185.2
|
TEX15
|
testis expressed 15, meiosis and synapsis associated |
| chrX_-_129843806 | 0.32 |
ENST00000357166.11
|
ZDHHC9
|
zinc finger DHHC-type palmitoyltransferase 9 |
| chr1_-_85708382 | 0.30 |
ENST00000370574.4
ENST00000431532.6 |
ZNHIT6
|
zinc finger HIT-type containing 6 |
| chr1_+_218346235 | 0.30 |
ENST00000366929.4
|
TGFB2
|
transforming growth factor beta 2 |
| chr15_+_58431985 | 0.30 |
ENST00000433326.2
ENST00000299022.10 |
LIPC
|
lipase C, hepatic type |
| chr11_-_57514876 | 0.29 |
ENST00000528450.5
|
SLC43A1
|
solute carrier family 43 member 1 |
| chr3_-_179071432 | 0.29 |
ENST00000414084.1
|
ZMAT3
|
zinc finger matrin-type 3 |
| chr6_+_13615322 | 0.29 |
ENST00000451315.7
|
NOL7
|
nucleolar protein 7 |
| chr17_-_54968697 | 0.28 |
ENST00000571584.1
|
COX11
|
cytochrome c oxidase copper chaperone COX11 |
| chr4_+_56436233 | 0.28 |
ENST00000512576.3
|
PAICS
|
phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase |
| chr2_-_135985568 | 0.27 |
ENST00000264161.9
|
DARS1
|
aspartyl-tRNA synthetase 1 |
| chr22_+_41301514 | 0.27 |
ENST00000352645.5
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr18_+_58196736 | 0.26 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr5_-_72320217 | 0.25 |
ENST00000508863.6
ENST00000522095.1 ENST00000513900.5 ENST00000515404.5 ENST00000261413.10 ENST00000457646.8 |
MRPS27
|
mitochondrial ribosomal protein S27 |
| chr16_+_19718264 | 0.24 |
ENST00000564186.5
|
IQCK
|
IQ motif containing K |
| chr1_+_20139316 | 0.23 |
ENST00000375102.4
|
PLA2G2F
|
phospholipase A2 group IIF |
| chr4_+_56435730 | 0.23 |
ENST00000514888.5
ENST00000264221.6 ENST00000505164.5 |
PAICS
|
phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase |
| chrX_-_129843388 | 0.23 |
ENST00000371064.7
|
ZDHHC9
|
zinc finger DHHC-type palmitoyltransferase 9 |
| chr6_-_154356735 | 0.23 |
ENST00000367220.8
ENST00000265198.8 ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr12_+_123762188 | 0.22 |
ENST00000409039.8
ENST00000673944.1 |
DNAH10
|
dynein axonemal heavy chain 10 |
| chr14_+_72926377 | 0.22 |
ENST00000353777.7
ENST00000358377.7 ENST00000394234.6 ENST00000509153.5 ENST00000555042.5 |
DCAF4
|
DDB1 and CUL4 associated factor 4 |
| chrX_+_55075023 | 0.22 |
ENST00000374971.2
|
PAGE2B
|
PAGE family member 2B |
| chr4_+_41981745 | 0.20 |
ENST00000333141.7
|
DCAF4L1
|
DDB1 and CUL4 associated factor 4 like 1 |
| chr22_-_24005409 | 0.20 |
ENST00000621179.5
|
GSTT4
|
glutathione S-transferase theta 4 |
| chr2_+_189784435 | 0.20 |
ENST00000409985.5
ENST00000441310.7 ENST00000446877.5 ENST00000409823.7 ENST00000374826.8 ENST00000424766.5 ENST00000447232.6 |
PMS1
|
PMS1 homolog 1, mismatch repair system component |
| chrX_+_55075062 | 0.20 |
ENST00000374974.7
|
PAGE2B
|
PAGE family member 2B |
| chr3_+_174859315 | 0.20 |
ENST00000454872.6
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase like 2 |
| chr15_-_74695987 | 0.20 |
ENST00000563009.5
ENST00000568176.5 ENST00000566243.5 ENST00000566219.1 ENST00000426797.7 ENST00000315127.9 ENST00000566119.5 |
EDC3
|
enhancer of mRNA decapping 3 |
| chr5_-_145835330 | 0.19 |
ENST00000394450.6
ENST00000683046.1 |
PRELID2
|
PRELI domain containing 2 |
| chr16_-_4847265 | 0.19 |
ENST00000591451.5
ENST00000436648.9 ENST00000321919.14 ENST00000588297.5 |
GLYR1
|
glyoxylate reductase 1 homolog |
| chr4_-_56435581 | 0.19 |
ENST00000264220.6
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase |
| chr5_+_90474879 | 0.19 |
ENST00000504930.5
ENST00000514483.5 |
POLR3G
|
RNA polymerase III subunit G |
| chr19_-_45406327 | 0.18 |
ENST00000593226.5
ENST00000418234.6 |
PPP1R13L
|
protein phosphatase 1 regulatory subunit 13 like |
| chr19_+_53554819 | 0.18 |
ENST00000505949.5
ENST00000513265.5 ENST00000648511.1 |
ZNF331
|
zinc finger protein 331 |
| chr12_-_54648356 | 0.18 |
ENST00000293371.11
ENST00000456047.2 |
DCD
|
dermcidin |
| chr11_-_56491370 | 0.18 |
ENST00000327216.5
|
OR5M8
|
olfactory receptor family 5 subfamily M member 8 |
| chr2_-_27323006 | 0.17 |
ENST00000402310.5
ENST00000405983.5 ENST00000403262.6 |
MPV17
|
mitochondrial inner membrane protein MPV17 |
| chr1_+_173824626 | 0.16 |
ENST00000648960.1
ENST00000648807.1 ENST00000649067.1 ENST00000649689.2 |
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr7_-_920884 | 0.16 |
ENST00000617043.4
ENST00000437486.5 |
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr2_+_206765578 | 0.16 |
ENST00000403094.3
ENST00000402774.8 |
FASTKD2
|
FAST kinase domains 2 |
| chr17_+_54968735 | 0.16 |
ENST00000376352.6
ENST00000405898.5 ENST00000434978.6 ENST00000398391.6 |
STXBP4
|
syntaxin binding protein 4 |
| chr1_-_200620729 | 0.15 |
ENST00000367350.5
|
KIF14
|
kinesin family member 14 |
| chr10_+_48306698 | 0.15 |
ENST00000374179.8
|
MAPK8
|
mitogen-activated protein kinase 8 |
| chr19_+_17933001 | 0.15 |
ENST00000445755.7
|
CCDC124
|
coiled-coil domain containing 124 |
| chr3_-_48301577 | 0.14 |
ENST00000451657.6
|
NME6
|
NME/NM23 nucleoside diphosphate kinase 6 |
| chr16_-_56668034 | 0.13 |
ENST00000569500.5
ENST00000379811.4 ENST00000444837.6 |
MT1G
|
metallothionein 1G |
| chr15_+_77420880 | 0.13 |
ENST00000336216.9
ENST00000558176.1 |
HMG20A
|
high mobility group 20A |
| chr22_-_37849296 | 0.13 |
ENST00000609454.5
|
ANKRD54
|
ankyrin repeat domain 54 |
| chr1_+_173824694 | 0.13 |
ENST00000647645.1
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr5_+_420914 | 0.13 |
ENST00000506456.1
|
AHRR
|
aryl-hydrocarbon receptor repressor |
| chrX_+_129906118 | 0.13 |
ENST00000425117.6
|
UTP14A
|
UTP14A small subunit processome component |
| chr17_-_8079903 | 0.12 |
ENST00000649809.1
|
ALOX12B
|
arachidonate 12-lipoxygenase, 12R type |
| chr19_+_53554499 | 0.12 |
ENST00000512387.6
ENST00000511567.5 |
ZNF331
|
zinc finger protein 331 |
| chr17_+_50719565 | 0.12 |
ENST00000625349.2
ENST00000393227.6 ENST00000240304.5 ENST00000505658.6 ENST00000505619.5 ENST00000510984.5 |
LUC7L3
|
LUC7 like 3 pre-mRNA splicing factor |
| chr6_-_36442843 | 0.11 |
ENST00000454782.3
|
PXT1
|
peroxisomal testis enriched protein 1 |
| chr22_-_43015133 | 0.11 |
ENST00000453643.5
ENST00000263246.8 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr4_+_15681683 | 0.11 |
ENST00000504137.1
|
FAM200B
|
family with sequence similarity 200 member B |
| chr1_+_34782259 | 0.11 |
ENST00000373362.3
|
GJB3
|
gap junction protein beta 3 |
| chr17_-_10697501 | 0.11 |
ENST00000577427.1
ENST00000255390.10 |
SCO1
|
synthesis of cytochrome C oxidase 1 |
| chr19_-_695493 | 0.11 |
ENST00000613411.4
|
PRSS57
|
serine protease 57 |
| chr10_+_23192306 | 0.10 |
ENST00000376504.4
|
PTF1A
|
pancreas associated transcription factor 1a |
| chr1_-_28643005 | 0.10 |
ENST00000263974.4
ENST00000373824.9 ENST00000495422.2 |
TAF12
|
TATA-box binding protein associated factor 12 |
| chr11_+_6603740 | 0.10 |
ENST00000537806.5
ENST00000420936.6 ENST00000299421.9 ENST00000528995.5 ENST00000396751.6 |
ILK
|
integrin linked kinase |
| chr17_-_8758559 | 0.10 |
ENST00000328794.10
|
SPDYE4
|
speedy/RINGO cell cycle regulator family member E4 |
| chr19_-_695427 | 0.09 |
ENST00000329267.9
|
PRSS57
|
serine protease 57 |
| chr6_+_143843316 | 0.09 |
ENST00000367576.6
|
LTV1
|
LTV1 ribosome biogenesis factor |
| chr12_+_93467506 | 0.09 |
ENST00000549982.6
ENST00000552217.6 ENST00000393128.8 ENST00000547098.5 |
MRPL42
|
mitochondrial ribosomal protein L42 |
| chr9_+_127707015 | 0.09 |
ENST00000614677.1
|
CFAP157
|
cilia and flagella associated protein 157 |
| chr15_+_77420957 | 0.08 |
ENST00000559099.5
|
HMG20A
|
high mobility group 20A |
| chr17_+_36949285 | 0.08 |
ENST00000681062.1
ENST00000679881.1 ENST00000680782.1 |
AATF
|
apoptosis antagonizing transcription factor |
| chr6_+_167271163 | 0.07 |
ENST00000503433.5
|
UNC93A
|
unc-93 homolog A |
| chr6_+_44247866 | 0.07 |
ENST00000371554.2
|
HSP90AB1
|
heat shock protein 90 alpha family class B member 1 |
| chr11_+_6603708 | 0.07 |
ENST00000532063.5
|
ILK
|
integrin linked kinase |
| chr4_-_158173042 | 0.07 |
ENST00000592057.1
ENST00000393807.9 |
GASK1B
|
golgi associated kinase 1B |
| chr11_-_3126605 | 0.06 |
ENST00000525498.5
|
OSBPL5
|
oxysterol binding protein like 5 |
| chr7_-_100896123 | 0.06 |
ENST00000428317.7
|
ACHE
|
acetylcholinesterase (Cartwright blood group) |
| chr2_-_111884117 | 0.06 |
ENST00000341068.8
|
ANAPC1
|
anaphase promoting complex subunit 1 |
| chr5_+_36151989 | 0.06 |
ENST00000274254.9
|
SKP2
|
S-phase kinase associated protein 2 |
| chr2_-_46915745 | 0.05 |
ENST00000649435.1
ENST00000409105.5 ENST00000319466.9 ENST00000409973.5 ENST00000409913.5 |
MCFD2
|
multiple coagulation factor deficiency 2, ER cargo receptor complex subunit |
| chr21_+_36699100 | 0.05 |
ENST00000290399.11
|
SIM2
|
SIM bHLH transcription factor 2 |
| chr2_-_46916020 | 0.05 |
ENST00000409800.5
ENST00000409218.5 |
MCFD2
|
multiple coagulation factor deficiency 2, ER cargo receptor complex subunit |
| chr9_+_129139382 | 0.05 |
ENST00000423100.5
ENST00000674648.1 |
PTPA
ENSG00000235007.3
|
protein phosphatase 2 phosphatase activator novel protein |
| chr1_+_231241195 | 0.05 |
ENST00000436239.5
ENST00000366647.9 ENST00000416000.1 |
GNPAT
|
glyceronephosphate O-acyltransferase |
| chr5_-_90474765 | 0.05 |
ENST00000316610.7
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr4_+_88007624 | 0.04 |
ENST00000237596.7
|
PKD2
|
polycystin 2, transient receptor potential cation channel |
| chr9_+_127706975 | 0.04 |
ENST00000373295.7
|
CFAP157
|
cilia and flagella associated protein 157 |
| chr19_+_45406630 | 0.03 |
ENST00000309424.8
|
POLR1G
|
RNA polymerase I subunit G |
| chr7_-_6272575 | 0.02 |
ENST00000350796.8
|
CYTH3
|
cytohesin 3 |
| chrX_+_129906146 | 0.02 |
ENST00000394422.8
|
UTP14A
|
UTP14A small subunit processome component |
| chr7_-_6272639 | 0.02 |
ENST00000396741.3
|
CYTH3
|
cytohesin 3 |
| chr10_+_99732211 | 0.02 |
ENST00000370476.10
ENST00000370472.4 |
CUTC
|
cutC copper transporter |
| chr12_-_42144675 | 0.02 |
ENST00000280876.6
|
GXYLT1
|
glucoside xylosyltransferase 1 |
| chr15_-_74695936 | 0.01 |
ENST00000647659.1
ENST00000566828.5 |
EDC3
|
enhancer of mRNA decapping 3 |
| chr14_-_55027045 | 0.01 |
ENST00000455555.1
ENST00000360586.8 ENST00000420358.2 |
WDHD1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr11_-_61792581 | 0.01 |
ENST00000537328.6
|
TMEM258
|
transmembrane protein 258 |
| chr8_+_98117285 | 0.01 |
ENST00000401707.7
ENST00000522319.5 |
POP1
|
POP1 homolog, ribonuclease P/MRP subunit |
| chr3_-_51499950 | 0.01 |
ENST00000423656.5
ENST00000504652.5 ENST00000684031.1 |
DCAF1
|
DDB1 and CUL4 associated factor 1 |
| chr22_-_30591850 | 0.01 |
ENST00000335214.8
ENST00000406208.7 ENST00000402284.7 ENST00000354694.12 |
PES1
|
pescadillo ribosomal biogenesis factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BHLHE40
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.4 | 1.3 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.2 | 0.9 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 | 0.6 | GO:0030719 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.2 | 0.5 | GO:0046709 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.1 | 0.7 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.4 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 0.9 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.5 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.3 | GO:1905006 | negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.1 | 0.3 | GO:0070145 | mitochondrial asparaginyl-tRNA aminoacylation(GO:0070145) |
| 0.1 | 2.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 1.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.4 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.8 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.1 | 1.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.6 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.6 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.4 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.1 | 0.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.5 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.9 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.6 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.5 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 2.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.3 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.8 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.4 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.5 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) |
| 0.0 | 0.1 | GO:1901389 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.0 | 0.5 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 1.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.3 | GO:0034638 | very-low-density lipoprotein particle remodeling(GO:0034372) phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 1.2 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 1.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.8 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 2.3 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.7 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.1 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.5 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.1 | 0.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.5 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 1.0 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.1 | GO:1990913 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 0.0 | 1.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 2.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0070362 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.3 | 0.9 | GO:0004639 | phosphoribosylaminoimidazole carboxylase activity(GO:0004638) phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.2 | 0.6 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.1 | 0.7 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 2.2 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 0.5 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.3 | GO:0050560 | aspartate-tRNA ligase activity(GO:0004815) aspartate-tRNA(Asn) ligase activity(GO:0050560) |
| 0.1 | 0.5 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.5 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.5 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.8 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.8 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.4 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.1 | 0.8 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.4 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.7 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 1.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.6 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 0.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 1.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.4 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.8 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.8 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.8 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |