Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
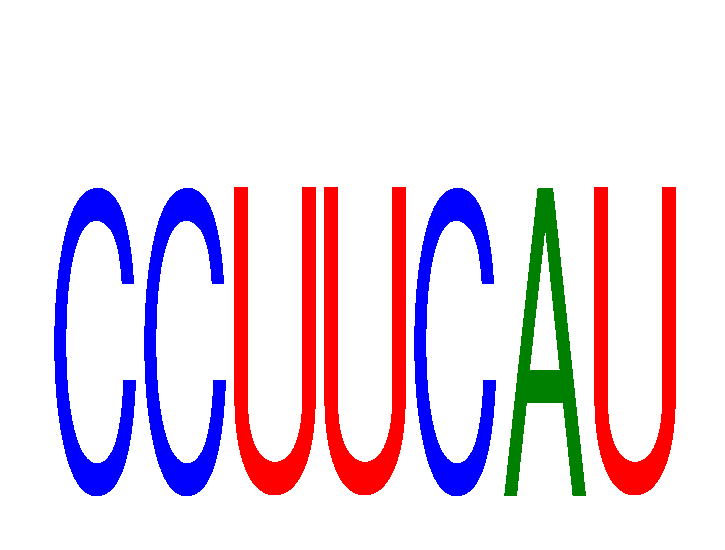
Results for CCUUCAU
Z-value: 0.64
miRNA associated with seed CCUUCAU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-205-5p
|
MIMAT0000266 |
Activity profile of CCUUCAU motif
Sorted Z-values of CCUUCAU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_168291599 | 2.61 |
ENST00000265293.9
|
WWC1
|
WW and C2 domain containing 1 |
| chr16_+_86510507 | 2.56 |
ENST00000262426.6
|
FOXF1
|
forkhead box F1 |
| chr2_-_187448244 | 1.63 |
ENST00000392370.8
ENST00000410068.5 ENST00000447403.5 ENST00000410102.5 |
CALCRL
|
calcitonin receptor like receptor |
| chr11_-_115504389 | 1.31 |
ENST00000545380.5
ENST00000452722.7 ENST00000331581.11 ENST00000537058.5 ENST00000536727.5 ENST00000542447.6 |
CADM1
|
cell adhesion molecule 1 |
| chr3_-_64687613 | 1.30 |
ENST00000295903.8
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif 9 |
| chr12_+_56080155 | 1.20 |
ENST00000267101.8
|
ERBB3
|
erb-b2 receptor tyrosine kinase 3 |
| chr16_+_4371840 | 1.01 |
ENST00000304735.4
|
VASN
|
vasorin |
| chr16_+_50548387 | 0.87 |
ENST00000268459.6
|
NKD1
|
NKD inhibitor of WNT signaling pathway 1 |
| chr4_+_76435216 | 0.82 |
ENST00000296043.7
|
SHROOM3
|
shroom family member 3 |
| chr10_+_58512864 | 0.82 |
ENST00000373886.8
|
BICC1
|
BicC family RNA binding protein 1 |
| chr4_+_71339014 | 0.74 |
ENST00000340595.4
|
SLC4A4
|
solute carrier family 4 member 4 |
| chr16_-_57479745 | 0.71 |
ENST00000566936.5
ENST00000568617.5 ENST00000567276.5 ENST00000569548.5 ENST00000569250.5 ENST00000564378.5 |
DOK4
|
docking protein 4 |
| chr6_-_8435480 | 0.70 |
ENST00000379660.4
|
SLC35B3
|
solute carrier family 35 member B3 |
| chr6_-_116060859 | 0.62 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr2_-_70553638 | 0.61 |
ENST00000444975.5
ENST00000445399.5 ENST00000295400.11 ENST00000418333.6 |
TGFA
|
transforming growth factor alpha |
| chr2_+_203328378 | 0.57 |
ENST00000430418.5
ENST00000261018.12 ENST00000295851.10 ENST00000424558.5 ENST00000417864.5 ENST00000422511.6 |
ABI2
|
abl interactor 2 |
| chr6_+_43076262 | 0.56 |
ENST00000476760.1
ENST00000230419.9 ENST00000471863.5 ENST00000345201.6 ENST00000349241.6 ENST00000352931.6 |
PTK7
|
protein tyrosine kinase 7 (inactive) |
| chr2_-_160493799 | 0.55 |
ENST00000348849.8
|
RBMS1
|
RNA binding motif single stranded interacting protein 1 |
| chr3_+_113838772 | 0.54 |
ENST00000358160.9
|
GRAMD1C
|
GRAM domain containing 1C |
| chr2_-_85561481 | 0.52 |
ENST00000233838.9
|
GGCX
|
gamma-glutamyl carboxylase |
| chr11_+_14643782 | 0.52 |
ENST00000282096.9
|
PDE3B
|
phosphodiesterase 3B |
| chr8_-_126558461 | 0.50 |
ENST00000304916.4
|
LRATD2
|
LRAT domain containing 2 |
| chr4_-_121072519 | 0.50 |
ENST00000379692.9
|
NDNF
|
neuron derived neurotrophic factor |
| chrX_-_129843806 | 0.49 |
ENST00000357166.11
|
ZDHHC9
|
zinc finger DHHC-type palmitoyltransferase 9 |
| chr10_+_28677487 | 0.45 |
ENST00000375533.6
|
BAMBI
|
BMP and activin membrane bound inhibitor |
| chr9_-_109320949 | 0.44 |
ENST00000374557.4
|
EPB41L4B
|
erythrocyte membrane protein band 4.1 like 4B |
| chr5_-_1523900 | 0.44 |
ENST00000283415.4
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1 |
| chr4_-_11428868 | 0.41 |
ENST00000002596.6
|
HS3ST1
|
heparan sulfate-glucosamine 3-sulfotransferase 1 |
| chr4_+_183099244 | 0.40 |
ENST00000403733.8
|
WWC2
|
WW and C2 domain containing 2 |
| chr3_+_150603279 | 0.40 |
ENST00000615547.4
ENST00000477889.5 ENST00000471696.6 ENST00000485923.1 |
SELENOT
|
selenoprotein T |
| chr15_+_69298896 | 0.39 |
ENST00000395407.7
ENST00000558684.5 |
PAQR5
|
progestin and adipoQ receptor family member 5 |
| chr3_+_170418856 | 0.37 |
ENST00000064724.8
ENST00000486975.1 |
CLDN11
ENSG00000285218.1
|
claudin 11 novel protein |
| chr1_+_109910840 | 0.36 |
ENST00000329608.11
ENST00000488198.5 |
CSF1
|
colony stimulating factor 1 |
| chr16_-_10580577 | 0.36 |
ENST00000359543.8
|
EMP2
|
epithelial membrane protein 2 |
| chr16_-_66696680 | 0.35 |
ENST00000330687.8
ENST00000563952.1 ENST00000394106.7 |
CMTM4
|
CKLF like MARVEL transmembrane domain containing 4 |
| chr3_-_116445458 | 0.34 |
ENST00000490035.7
|
LSAMP
|
limbic system associated membrane protein |
| chr3_+_61561561 | 0.34 |
ENST00000474889.6
|
PTPRG
|
protein tyrosine phosphatase receptor type G |
| chr11_+_102110437 | 0.33 |
ENST00000282441.10
ENST00000526343.5 ENST00000537274.5 ENST00000345877.6 ENST00000615667.4 |
YAP1
|
Yes1 associated transcriptional regulator |
| chr1_-_63523175 | 0.33 |
ENST00000371092.7
ENST00000271002.15 |
ITGB3BP
|
integrin subunit beta 3 binding protein |
| chr16_-_65121930 | 0.33 |
ENST00000566827.5
ENST00000394156.7 ENST00000268603.9 ENST00000562998.1 |
CDH11
|
cadherin 11 |
| chr13_+_42781547 | 0.32 |
ENST00000537894.5
|
FAM216B
|
family with sequence similarity 216 member B |
| chr10_+_8054668 | 0.32 |
ENST00000379328.9
|
GATA3
|
GATA binding protein 3 |
| chr6_+_43770707 | 0.32 |
ENST00000324450.11
ENST00000417285.7 ENST00000413642.8 ENST00000372055.9 ENST00000482630.7 ENST00000425836.7 ENST00000372064.9 ENST00000372077.8 ENST00000519767.5 |
VEGFA
|
vascular endothelial growth factor A |
| chr8_-_37899454 | 0.32 |
ENST00000522727.5
ENST00000287263.8 ENST00000330843.9 |
RAB11FIP1
|
RAB11 family interacting protein 1 |
| chr1_-_185317234 | 0.31 |
ENST00000367498.8
|
IVNS1ABP
|
influenza virus NS1A binding protein |
| chr3_+_160225409 | 0.31 |
ENST00000326474.5
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr12_+_22625075 | 0.31 |
ENST00000671733.1
ENST00000335148.8 ENST00000672951.1 ENST00000266517.9 |
ETNK1
|
ethanolamine kinase 1 |
| chr2_-_175005357 | 0.30 |
ENST00000409156.7
ENST00000444573.2 ENST00000409900.9 |
CHN1
|
chimerin 1 |
| chr15_+_39581068 | 0.30 |
ENST00000397591.2
ENST00000260356.6 |
THBS1
|
thrombospondin 1 |
| chr10_-_96586975 | 0.28 |
ENST00000371142.9
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr17_-_64506281 | 0.28 |
ENST00000225792.10
ENST00000585060.5 |
DDX5
|
DEAD-box helicase 5 |
| chr3_+_152835122 | 0.28 |
ENST00000305097.6
|
P2RY1
|
purinergic receptor P2Y1 |
| chr13_-_61415508 | 0.27 |
ENST00000409204.4
|
PCDH20
|
protocadherin 20 |
| chr1_+_113390495 | 0.26 |
ENST00000307546.14
|
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr20_+_50731571 | 0.26 |
ENST00000371610.7
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr2_-_127643212 | 0.26 |
ENST00000409286.5
|
LIMS2
|
LIM zinc finger domain containing 2 |
| chr1_+_183023409 | 0.25 |
ENST00000258341.5
|
LAMC1
|
laminin subunit gamma 1 |
| chr11_+_120325283 | 0.25 |
ENST00000314475.6
ENST00000375095.3 ENST00000529187.1 |
TLCD5
|
TLC domain containing 5 |
| chr7_-_41703062 | 0.25 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr1_+_213987929 | 0.24 |
ENST00000498508.6
ENST00000366958.9 |
PROX1
|
prospero homeobox 1 |
| chr1_+_201955496 | 0.24 |
ENST00000367287.5
|
TIMM17A
|
translocase of inner mitochondrial membrane 17A |
| chr6_-_31862809 | 0.24 |
ENST00000375631.5
|
NEU1
|
neuraminidase 1 |
| chr16_+_69565958 | 0.23 |
ENST00000349945.7
ENST00000354436.6 |
NFAT5
|
nuclear factor of activated T cells 5 |
| chr4_-_39638846 | 0.23 |
ENST00000295958.10
|
SMIM14
|
small integral membrane protein 14 |
| chr18_-_50195138 | 0.23 |
ENST00000285039.12
|
MYO5B
|
myosin VB |
| chr8_+_17027230 | 0.21 |
ENST00000318063.10
|
MICU3
|
mitochondrial calcium uptake family member 3 |
| chr4_-_87391149 | 0.21 |
ENST00000507286.1
ENST00000358290.9 |
HSD17B11
|
hydroxysteroid 17-beta dehydrogenase 11 |
| chr20_+_11890723 | 0.20 |
ENST00000254977.7
|
BTBD3
|
BTB domain containing 3 |
| chr1_+_107056656 | 0.20 |
ENST00000370078.2
|
PRMT6
|
protein arginine methyltransferase 6 |
| chr4_+_76949743 | 0.20 |
ENST00000502584.5
ENST00000264893.11 ENST00000510641.5 |
SEPTIN11
|
septin 11 |
| chr12_-_62935117 | 0.19 |
ENST00000228705.7
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent 1H |
| chr4_-_5893075 | 0.19 |
ENST00000324989.12
|
CRMP1
|
collapsin response mediator protein 1 |
| chr12_+_40224956 | 0.19 |
ENST00000298910.12
ENST00000343742.6 |
LRRK2
|
leucine rich repeat kinase 2 |
| chr15_-_74433942 | 0.19 |
ENST00000543145.6
ENST00000261918.9 |
SEMA7A
|
semaphorin 7A (John Milton Hagen blood group) |
| chr9_-_10612966 | 0.19 |
ENST00000381196.9
|
PTPRD
|
protein tyrosine phosphatase receptor type D |
| chr11_-_86069043 | 0.19 |
ENST00000532317.5
ENST00000528256.1 ENST00000393346.8 ENST00000526033.5 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr2_+_176122712 | 0.18 |
ENST00000249499.8
|
HOXD9
|
homeobox D9 |
| chr12_-_114684151 | 0.18 |
ENST00000349155.7
|
TBX3
|
T-box transcription factor 3 |
| chr1_+_180632001 | 0.18 |
ENST00000367590.9
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr17_+_7035979 | 0.18 |
ENST00000308027.7
|
SLC16A13
|
solute carrier family 16 member 13 |
| chr15_-_72197772 | 0.18 |
ENST00000309731.12
|
GRAMD2A
|
GRAM domain containing 2A |
| chr8_+_69466617 | 0.17 |
ENST00000525061.5
ENST00000260128.8 ENST00000458141.6 |
SULF1
|
sulfatase 1 |
| chr4_-_41214602 | 0.17 |
ENST00000508676.5
ENST00000506352.5 ENST00000295974.12 |
APBB2
|
amyloid beta precursor protein binding family B member 2 |
| chr2_+_45651650 | 0.16 |
ENST00000306156.8
|
PRKCE
|
protein kinase C epsilon |
| chr12_-_12267003 | 0.16 |
ENST00000535731.1
ENST00000261349.9 |
LRP6
|
LDL receptor related protein 6 |
| chr6_+_87472925 | 0.16 |
ENST00000369556.7
ENST00000369557.9 ENST00000369552.9 |
SLC35A1
|
solute carrier family 35 member A1 |
| chr20_+_36092698 | 0.16 |
ENST00000430276.5
ENST00000373950.6 ENST00000373946.7 ENST00000441639.5 ENST00000628415.2 ENST00000452261.5 |
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr18_-_31684504 | 0.15 |
ENST00000383131.3
ENST00000237019.11 ENST00000306851.10 |
B4GALT6
|
beta-1,4-galactosyltransferase 6 |
| chr4_+_15002443 | 0.15 |
ENST00000538197.7
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr21_-_14383125 | 0.15 |
ENST00000285667.4
|
HSPA13
|
heat shock protein family A (Hsp70) member 13 |
| chr2_-_207166818 | 0.15 |
ENST00000423015.5
|
KLF7
|
Kruppel like factor 7 |
| chr1_+_26280117 | 0.15 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamate rich protein like 3 |
| chr1_-_23980308 | 0.15 |
ENST00000374452.9
ENST00000492112.3 ENST00000343255.9 ENST00000344989.10 |
SRSF10
|
serine and arginine rich splicing factor 10 |
| chr6_+_36954729 | 0.14 |
ENST00000373674.4
|
PI16
|
peptidase inhibitor 16 |
| chr12_+_56521798 | 0.13 |
ENST00000262031.10
|
RBMS2
|
RNA binding motif single stranded interacting protein 2 |
| chr6_-_110815408 | 0.13 |
ENST00000368911.8
|
CDK19
|
cyclin dependent kinase 19 |
| chr4_-_82798735 | 0.13 |
ENST00000273908.4
ENST00000319540.9 |
SCD5
|
stearoyl-CoA desaturase 5 |
| chr14_+_36661852 | 0.13 |
ENST00000361487.7
|
PAX9
|
paired box 9 |
| chr8_-_98825628 | 0.13 |
ENST00000617590.1
ENST00000518165.5 ENST00000419617.7 |
STK3
|
serine/threonine kinase 3 |
| chr2_-_40452046 | 0.12 |
ENST00000406785.6
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr2_-_96870034 | 0.12 |
ENST00000305476.10
|
SEMA4C
|
semaphorin 4C |
| chr14_-_77028663 | 0.12 |
ENST00000238647.5
|
IRF2BPL
|
interferon regulatory factor 2 binding protein like |
| chr9_+_33025265 | 0.12 |
ENST00000330899.5
|
DNAJA1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr4_-_98929092 | 0.11 |
ENST00000280892.10
ENST00000511644.5 ENST00000504432.5 ENST00000450253.7 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr1_+_70411180 | 0.11 |
ENST00000411986.6
|
CTH
|
cystathionine gamma-lyase |
| chr5_-_132490750 | 0.11 |
ENST00000437654.6
ENST00000245414.9 ENST00000680139.1 ENST00000680352.1 ENST00000679440.1 ENST00000680903.1 |
IRF1
|
interferon regulatory factor 1 |
| chr18_-_26090584 | 0.11 |
ENST00000415083.7
|
SS18
|
SS18 subunit of BAF chromatin remodeling complex |
| chr4_+_74365136 | 0.11 |
ENST00000244869.3
|
EREG
|
epiregulin |
| chrX_+_69504320 | 0.10 |
ENST00000252338.5
|
FAM155B
|
family with sequence similarity 155 member B |
| chr7_-_79453544 | 0.10 |
ENST00000419488.5
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr4_-_124712721 | 0.10 |
ENST00000504087.6
ENST00000515641.1 |
ANKRD50
|
ankyrin repeat domain 50 |
| chr12_-_54419259 | 0.10 |
ENST00000293379.9
|
ITGA5
|
integrin subunit alpha 5 |
| chr8_-_94949350 | 0.10 |
ENST00000448464.6
ENST00000342697.5 |
TP53INP1
|
tumor protein p53 inducible nuclear protein 1 |
| chr7_-_108456378 | 0.10 |
ENST00000613830.4
ENST00000413765.6 ENST00000379028.8 |
NRCAM
|
neuronal cell adhesion molecule |
| chr14_-_70416994 | 0.09 |
ENST00000621525.4
ENST00000256366.6 |
SYNJ2BP-COX16
SYNJ2BP
|
SYNJ2BP-COX16 readthrough synaptojanin 2 binding protein |
| chr15_+_58771280 | 0.09 |
ENST00000559228.6
ENST00000450403.3 |
MINDY2
|
MINDY lysine 48 deubiquitinase 2 |
| chr22_-_37486357 | 0.09 |
ENST00000356998.8
ENST00000416983.7 ENST00000424765.2 |
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_-_155241220 | 0.09 |
ENST00000368373.8
ENST00000427500.7 |
GBA
|
glucosylceramidase beta |
| chr14_-_70809494 | 0.08 |
ENST00000381250.8
ENST00000554752.7 ENST00000555993.6 |
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr2_+_176092715 | 0.08 |
ENST00000392539.4
|
HOXD13
|
homeobox D13 |
| chr5_+_116084992 | 0.08 |
ENST00000274458.9
ENST00000632434.1 |
COMMD10
|
COMM domain containing 10 |
| chr2_+_181985846 | 0.08 |
ENST00000682840.1
ENST00000409137.7 ENST00000280295.7 |
PPP1R1C
|
protein phosphatase 1 regulatory inhibitor subunit 1C |
| chr9_-_121201836 | 0.08 |
ENST00000373840.9
|
RAB14
|
RAB14, member RAS oncogene family |
| chr13_+_52652828 | 0.08 |
ENST00000310528.9
ENST00000343788.10 |
SUGT1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr17_-_49414871 | 0.08 |
ENST00000504124.6
ENST00000300408.8 ENST00000614445.5 |
PHB
|
prohibitin |
| chr1_+_93448155 | 0.08 |
ENST00000370253.6
|
FNBP1L
|
formin binding protein 1 like |
| chr10_+_95907965 | 0.07 |
ENST00000423344.6
ENST00000646931.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr1_+_100896060 | 0.07 |
ENST00000370112.8
ENST00000357650.9 |
SLC30A7
|
solute carrier family 30 member 7 |
| chr16_+_22008083 | 0.07 |
ENST00000542527.7
ENST00000569656.5 ENST00000562695.1 |
MOSMO
|
modulator of smoothened |
| chrX_+_23334841 | 0.07 |
ENST00000379361.5
|
PTCHD1
|
patched domain containing 1 |
| chr12_+_63844663 | 0.07 |
ENST00000355086.8
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1 |
| chr6_-_158818225 | 0.07 |
ENST00000337147.11
|
EZR
|
ezrin |
| chr19_+_13795434 | 0.07 |
ENST00000254323.6
|
ZSWIM4
|
zinc finger SWIM-type containing 4 |
| chr13_+_99501464 | 0.07 |
ENST00000376387.5
|
TM9SF2
|
transmembrane 9 superfamily member 2 |
| chr12_-_93441886 | 0.06 |
ENST00000552442.1
ENST00000550657.1 ENST00000318066.7 |
UBE2N
|
ubiquitin conjugating enzyme E2 N |
| chr12_+_70366277 | 0.06 |
ENST00000258111.5
|
KCNMB4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chr17_+_15999815 | 0.06 |
ENST00000261647.10
|
TTC19
|
tetratricopeptide repeat domain 19 |
| chr4_+_145481845 | 0.06 |
ENST00000302085.9
ENST00000512019.1 |
SMAD1
|
SMAD family member 1 |
| chr12_-_109573482 | 0.06 |
ENST00000540016.5
ENST00000545712.7 |
MMAB
|
metabolism of cobalamin associated B |
| chr21_+_32412648 | 0.06 |
ENST00000401402.7
ENST00000382699.7 ENST00000300255.7 |
EVA1C
|
eva-1 homolog C |
| chr7_+_128739292 | 0.06 |
ENST00000535011.6
ENST00000542996.6 ENST00000249364.9 ENST00000449187.6 |
CALU
|
calumenin |
| chr9_-_124771304 | 0.06 |
ENST00000416460.6
ENST00000487099.7 |
NR6A1
|
nuclear receptor subfamily 6 group A member 1 |
| chr2_-_100104530 | 0.05 |
ENST00000432037.5
ENST00000673232.1 ENST00000423966.6 ENST00000409236.6 |
AFF3
|
AF4/FMR2 family member 3 |
| chr10_+_68332055 | 0.05 |
ENST00000265866.12
|
HNRNPH3
|
heterogeneous nuclear ribonucleoprotein H3 |
| chr5_-_149551381 | 0.05 |
ENST00000670598.1
ENST00000657001.1 ENST00000515768.6 ENST00000261798.10 ENST00000377843.8 |
CSNK1A1
|
casein kinase 1 alpha 1 |
| chr1_-_217089627 | 0.05 |
ENST00000361525.7
|
ESRRG
|
estrogen related receptor gamma |
| chr2_+_149330506 | 0.05 |
ENST00000334166.9
|
LYPD6
|
LY6/PLAUR domain containing 6 |
| chr11_+_121024072 | 0.05 |
ENST00000529397.5
ENST00000683345.1 ENST00000422003.6 |
TBCEL
|
tubulin folding cofactor E like |
| chr2_-_201451446 | 0.05 |
ENST00000332624.8
ENST00000430254.1 |
TRAK2
|
trafficking kinesin protein 2 |
| chr11_-_22829793 | 0.05 |
ENST00000354193.5
|
SVIP
|
small VCP interacting protein |
| chr9_-_112175283 | 0.05 |
ENST00000374270.8
ENST00000374263.7 |
SUSD1
|
sushi domain containing 1 |
| chr8_+_55879818 | 0.05 |
ENST00000520220.6
ENST00000519728.6 |
LYN
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr3_+_141387801 | 0.05 |
ENST00000514251.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr10_+_59176600 | 0.05 |
ENST00000373880.9
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein like |
| chr7_+_90596281 | 0.05 |
ENST00000380050.8
|
CDK14
|
cyclin dependent kinase 14 |
| chr12_+_8082260 | 0.04 |
ENST00000638237.1
ENST00000339754.11 ENST00000639811.1 ENST00000639167.1 ENST00000541948.2 |
NECAP1
|
NECAP endocytosis associated 1 |
| chr10_+_123135938 | 0.04 |
ENST00000357878.7
|
HMX3
|
H6 family homeobox 3 |
| chrX_+_73563190 | 0.04 |
ENST00000373504.10
ENST00000373502.9 |
CHIC1
|
cysteine rich hydrophobic domain 1 |
| chr4_+_85475131 | 0.04 |
ENST00000395184.6
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chrX_+_10015226 | 0.04 |
ENST00000380861.9
|
WWC3
|
WWC family member 3 |
| chr1_+_101237009 | 0.04 |
ENST00000305352.7
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr12_+_130872037 | 0.04 |
ENST00000448750.7
ENST00000541630.5 ENST00000543796.6 ENST00000392369.6 ENST00000535090.5 ENST00000541679.7 ENST00000392367.4 |
RAN
|
RAN, member RAS oncogene family |
| chrX_-_20266834 | 0.03 |
ENST00000379565.9
|
RPS6KA3
|
ribosomal protein S6 kinase A3 |
| chr22_+_20080211 | 0.03 |
ENST00000383024.6
ENST00000351989.8 |
DGCR8
|
DGCR8 microprocessor complex subunit |
| chr2_+_24491860 | 0.03 |
ENST00000406961.5
ENST00000405141.5 |
NCOA1
|
nuclear receptor coactivator 1 |
| chr4_+_26860778 | 0.03 |
ENST00000467011.6
|
STIM2
|
stromal interaction molecule 2 |
| chr11_+_47980538 | 0.03 |
ENST00000613246.4
ENST00000418331.7 ENST00000615445.4 ENST00000440289.6 |
PTPRJ
|
protein tyrosine phosphatase receptor type J |
| chr16_-_3443446 | 0.03 |
ENST00000301744.7
|
ZNF597
|
zinc finger protein 597 |
| chr14_-_99604167 | 0.03 |
ENST00000380243.9
|
CCDC85C
|
coiled-coil domain containing 85C |
| chrX_-_103832204 | 0.03 |
ENST00000674363.1
ENST00000674162.1 ENST00000674338.1 ENST00000674274.1 ENST00000674271.1 ENST00000674265.1 ENST00000674212.1 ENST00000674255.1 ENST00000674342.1 ENST00000674430.1 ENST00000243298.3 |
ENSG00000288597.1
RAB9B
|
novel transcript RAB9B, member RAS oncogene family |
| chr20_-_33686371 | 0.03 |
ENST00000343380.6
|
E2F1
|
E2F transcription factor 1 |
| chr8_-_19013693 | 0.03 |
ENST00000327040.13
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr6_-_52577012 | 0.03 |
ENST00000182527.4
|
TRAM2
|
translocation associated membrane protein 2 |
| chr11_+_66975271 | 0.03 |
ENST00000308963.4
|
C11orf86
|
chromosome 11 open reading frame 86 |
| chr6_-_84764581 | 0.02 |
ENST00000369663.10
|
TBX18
|
T-box transcription factor 18 |
| chrX_+_77910656 | 0.02 |
ENST00000343533.9
ENST00000341514.11 ENST00000645454.1 ENST00000642651.1 ENST00000644362.1 |
ATP7A
PGK1
|
ATPase copper transporting alpha phosphoglycerate kinase 1 |
| chr2_+_28981295 | 0.02 |
ENST00000379558.5
|
TOGARAM2
|
TOG array regulator of axonemal microtubules 2 |
| chr12_+_53954870 | 0.02 |
ENST00000243103.4
|
HOXC12
|
homeobox C12 |
| chr1_+_192158448 | 0.02 |
ENST00000367460.4
|
RGS18
|
regulator of G protein signaling 18 |
| chr2_+_108449178 | 0.02 |
ENST00000309863.11
ENST00000409821.5 |
GCC2
|
GRIP and coiled-coil domain containing 2 |
| chr7_-_10940123 | 0.02 |
ENST00000339600.6
|
NDUFA4
|
NDUFA4 mitochondrial complex associated |
| chr12_+_96194365 | 0.02 |
ENST00000228741.8
ENST00000547249.1 |
ELK3
|
ETS transcription factor ELK3 |
| chr20_-_49482645 | 0.02 |
ENST00000371741.6
|
KCNB1
|
potassium voltage-gated channel subfamily B member 1 |
| chr4_-_16226460 | 0.01 |
ENST00000405303.7
|
TAPT1
|
transmembrane anterior posterior transformation 1 |
| chr10_+_17752185 | 0.01 |
ENST00000377495.2
|
TMEM236
|
transmembrane protein 236 |
| chr13_-_107867467 | 0.01 |
ENST00000375915.4
|
FAM155A
|
family with sequence similarity 155 member A |
| chr5_+_119071358 | 0.01 |
ENST00000311085.8
|
DMXL1
|
Dmx like 1 |
| chr12_+_19439469 | 0.01 |
ENST00000266508.14
|
AEBP2
|
AE binding protein 2 |
| chr12_+_111034136 | 0.01 |
ENST00000261726.11
|
CUX2
|
cut like homeobox 2 |
| chr1_+_167220870 | 0.01 |
ENST00000367866.7
ENST00000429375.6 ENST00000541643.7 |
POU2F1
|
POU class 2 homeobox 1 |
| chr10_+_99329349 | 0.01 |
ENST00000356713.5
|
CNNM1
|
cyclin and CBS domain divalent metal cation transport mediator 1 |
| chr2_+_138501753 | 0.01 |
ENST00000280098.9
|
SPOPL
|
speckle type BTB/POZ protein like |
| chr13_-_30464234 | 0.00 |
ENST00000399489.5
ENST00000339872.8 |
HMGB1
|
high mobility group box 1 |
| chr6_+_45328203 | 0.00 |
ENST00000371432.7
ENST00000647337.2 ENST00000371438.5 |
RUNX2
|
RUNX family transcription factor 2 |
| chr4_+_123399488 | 0.00 |
ENST00000394339.2
|
SPRY1
|
sprouty RTK signaling antagonist 1 |
| chr19_-_31349408 | 0.00 |
ENST00000240587.5
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr16_+_67846917 | 0.00 |
ENST00000219169.9
ENST00000567105.5 |
NUTF2
|
nuclear transport factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CCUUCAU
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0060458 | right lung development(GO:0060458) |
| 0.3 | 0.9 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 3.0 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 1.0 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 1.3 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.7 | GO:0046963 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.1 | 1.6 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.3 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.3 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.4 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 0.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.2 | GO:0043049 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 0.3 | GO:2000607 | regulation of cell proliferation involved in mesonephros development(GO:2000606) negative regulation of cell proliferation involved in mesonephros development(GO:2000607) fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000699) glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000701) regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000702) negative regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000703) regulation of glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000733) negative regulation of glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000734) |
| 0.1 | 0.4 | GO:1904141 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.1 | 0.2 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.2 | GO:2000469 | regulation of thioredoxin peroxidase activity(GO:1903123) negative regulation of thioredoxin peroxidase activity(GO:1903124) negative regulation of thioredoxin peroxidase activity by peptidyl-threonine phosphorylation(GO:1903125) Wnt signalosome assembly(GO:1904887) negative regulation of peroxidase activity(GO:2000469) |
| 0.1 | 0.2 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 1.3 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.1 | 0.2 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.1 | 0.2 | GO:0044335 | canonical Wnt signaling pathway involved in neural crest cell differentiation(GO:0044335) |
| 0.0 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.0 | 0.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.6 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.8 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.2 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.5 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.4 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.5 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.1 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.0 | 0.6 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.3 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.6 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.2 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 1.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.4 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:1901805 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.2 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.2 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.3 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.1 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.0 | 0.5 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0070662 | mast cell proliferation(GO:0070662) |
| 0.0 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.4 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.3 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.0 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.0 | 0.1 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.0 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.2 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.1 | 0.2 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.3 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.2 | GO:0099400 | cytoplasmic side of mitochondrial outer membrane(GO:0032473) caveola neck(GO:0099400) |
| 0.0 | 0.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.0 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.0 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.0 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 2.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.2 | 0.5 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.2 | 0.5 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.1 | 0.7 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.4 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 1.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.2 | GO:0036479 | GTP-dependent protein kinase activity(GO:0034211) peroxidase inhibitor activity(GO:0036479) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.3 | GO:0070051 | fibrinogen binding(GO:0070051) collagen V binding(GO:0070052) |
| 0.1 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.2 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.1 | 0.6 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.0 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.4 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.3 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.2 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 1.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.3 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.4 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 3.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 2.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 2.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.0 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |