Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
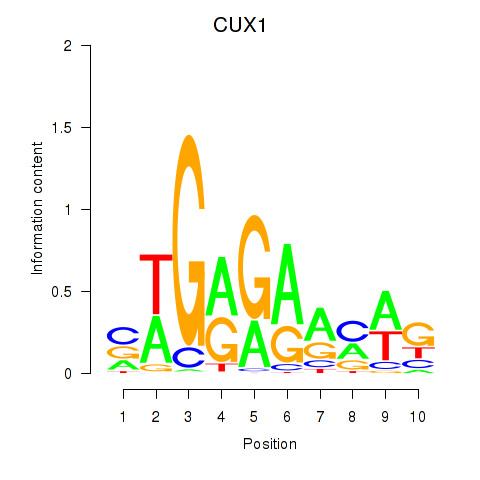
Results for CUX1
Z-value: 1.41
Transcription factors associated with CUX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CUX1
|
ENSG00000257923.12 | cut like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CUX1 | hg38_v1_chr7_+_101815983_101816030 | -0.41 | 1.9e-02 | Click! |
Activity profile of CUX1 motif
Sorted Z-values of CUX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_107719657 | 4.15 |
ENST00000525934.1
ENST00000531293.1 |
SLN
|
sarcolipin |
| chr19_-_55147319 | 2.93 |
ENST00000593046.5
|
TNNT1
|
troponin T1, slow skeletal type |
| chr17_-_76493133 | 2.82 |
ENST00000585701.5
ENST00000591192.1 ENST00000589526.5 |
RHBDF2
|
rhomboid 5 homolog 2 |
| chr19_-_55146894 | 2.81 |
ENST00000585321.6
ENST00000587465.6 |
TNNT1
|
troponin T1, slow skeletal type |
| chr12_-_110920568 | 2.61 |
ENST00000548438.1
ENST00000228841.15 |
MYL2
|
myosin light chain 2 |
| chr1_+_161706949 | 2.53 |
ENST00000350710.3
ENST00000367949.6 ENST00000367959.6 ENST00000540521.5 ENST00000546024.5 ENST00000674251.1 ENST00000674323.1 |
FCRLA
|
Fc receptor like A |
| chr13_-_113453524 | 2.42 |
ENST00000612156.2
ENST00000375418.7 |
ADPRHL1
|
ADP-ribosylhydrolase like 1 |
| chr22_+_22431949 | 2.38 |
ENST00000390301.3
|
IGLV1-36
|
immunoglobulin lambda variable 1-36 |
| chr15_-_34795541 | 2.33 |
ENST00000290378.6
|
ACTC1
|
actin alpha cardiac muscle 1 |
| chr1_+_161707244 | 2.28 |
ENST00000349527.8
ENST00000294796.8 ENST00000309691.10 ENST00000367953.7 ENST00000367950.2 |
FCRLA
|
Fc receptor like A |
| chr19_-_49155384 | 2.24 |
ENST00000252825.9
|
HRC
|
histidine rich calcium binding protein |
| chr19_-_45493208 | 2.15 |
ENST00000430715.6
|
RTN2
|
reticulon 2 |
| chr4_+_119135825 | 2.12 |
ENST00000307128.6
|
MYOZ2
|
myozenin 2 |
| chr5_+_150601060 | 2.08 |
ENST00000394243.5
|
SYNPO
|
synaptopodin |
| chr2_+_167135901 | 2.06 |
ENST00000628543.2
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr19_-_55147281 | 1.90 |
ENST00000589226.5
|
TNNT1
|
troponin T1, slow skeletal type |
| chr16_-_4242068 | 1.83 |
ENST00000399609.7
|
SRL
|
sarcalumenin |
| chr1_+_160115715 | 1.83 |
ENST00000361216.8
|
ATP1A2
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr2_-_157325808 | 1.77 |
ENST00000410096.6
ENST00000420719.6 ENST00000409216.5 ENST00000419116.2 |
ERMN
|
ermin |
| chr19_-_55140922 | 1.76 |
ENST00000589745.5
|
TNNT1
|
troponin T1, slow skeletal type |
| chr6_+_41053194 | 1.73 |
ENST00000244669.3
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme catalytic subunit 2 |
| chr1_-_154194648 | 1.70 |
ENST00000515609.1
|
TPM3
|
tropomyosin 3 |
| chr12_-_57237090 | 1.68 |
ENST00000556732.1
|
NDUFA4L2
|
NDUFA4 mitochondrial complex associated like 2 |
| chr1_+_153628393 | 1.66 |
ENST00000368696.3
ENST00000292169.6 ENST00000436839.1 |
S100A1
|
S100 calcium binding protein A1 |
| chr9_-_89498038 | 1.65 |
ENST00000339861.8
ENST00000422704.7 ENST00000455551.6 |
SEMA4D
|
semaphorin 4D |
| chr4_+_70637662 | 1.59 |
ENST00000472597.1
ENST00000472903.5 |
ENAM
ENSG00000286848.1
|
enamelin novel transcript |
| chr2_+_87338511 | 1.57 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr1_+_161707205 | 1.57 |
ENST00000367957.7
|
FCRLA
|
Fc receptor like A |
| chr1_-_16017825 | 1.56 |
ENST00000463576.5
|
HSPB7
|
heat shock protein family B (small) member 7 |
| chr19_-_7700074 | 1.55 |
ENST00000593418.1
|
FCER2
|
Fc fragment of IgE receptor II |
| chr12_+_85874287 | 1.54 |
ENST00000551529.5
ENST00000256010.7 |
NTS
|
neurotensin |
| chr3_+_159273235 | 1.52 |
ENST00000638749.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr1_+_161707222 | 1.52 |
ENST00000236938.12
|
FCRLA
|
Fc receptor like A |
| chr14_-_70080180 | 1.51 |
ENST00000394330.6
ENST00000533541.1 ENST00000216568.11 |
SLC8A3
|
solute carrier family 8 member A3 |
| chr5_-_139395096 | 1.50 |
ENST00000434752.4
|
PROB1
|
proline rich basic protein 1 |
| chr5_-_16617085 | 1.49 |
ENST00000684521.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr7_-_44225893 | 1.45 |
ENST00000425809.5
|
CAMK2B
|
calcium/calmodulin dependent protein kinase II beta |
| chr5_+_150640678 | 1.44 |
ENST00000519664.1
|
SYNPO
|
synaptopodin |
| chr1_+_160127672 | 1.43 |
ENST00000447527.1
|
ATP1A2
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr9_+_12693327 | 1.42 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
| chr14_-_106012390 | 1.41 |
ENST00000455737.2
|
IGHV4-4
|
immunoglobulin heavy variable 4-4 |
| chr17_+_50165990 | 1.40 |
ENST00000262018.8
ENST00000344627.10 ENST00000682109.1 ENST00000504073.2 |
SGCA
|
sarcoglycan alpha |
| chr10_-_75099489 | 1.39 |
ENST00000472493.6
ENST00000478873.7 ENST00000605915.5 |
DUSP13
|
dual specificity phosphatase 13 |
| chr3_-_39192584 | 1.39 |
ENST00000340369.4
ENST00000421646.1 ENST00000396251.1 |
XIRP1
|
xin actin binding repeat containing 1 |
| chr10_-_73641450 | 1.38 |
ENST00000359322.5
|
MYOZ1
|
myozenin 1 |
| chr15_-_42156590 | 1.33 |
ENST00000397272.7
|
PLA2G4F
|
phospholipase A2 group IVF |
| chr14_-_106185387 | 1.33 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18 |
| chr15_-_21718245 | 1.33 |
ENST00000630556.1
|
ENSG00000281179.1
|
novel gene identicle to IGHV1OR15-1 |
| chr1_+_63593354 | 1.32 |
ENST00000650546.1
ENST00000371084.8 |
PGM1
|
phosphoglucomutase 1 |
| chr19_-_49441508 | 1.32 |
ENST00000221485.8
|
SLC17A7
|
solute carrier family 17 member 7 |
| chr5_-_151347552 | 1.32 |
ENST00000335244.9
ENST00000521967.1 |
SLC36A2
|
solute carrier family 36 member 2 |
| chr5_-_176630517 | 1.31 |
ENST00000393693.7
ENST00000614675.4 |
SNCB
|
synuclein beta |
| chr9_-_127873462 | 1.30 |
ENST00000223836.10
|
AK1
|
adenylate kinase 1 |
| chr10_-_73433550 | 1.29 |
ENST00000299432.7
|
MSS51
|
MSS51 mitochondrial translational activator |
| chr5_-_16616972 | 1.28 |
ENST00000682564.1
ENST00000306320.10 ENST00000682229.1 |
RETREG1
|
reticulophagy regulator 1 |
| chr1_-_11847772 | 1.26 |
ENST00000376480.7
ENST00000610706.1 |
NPPA
|
natriuretic peptide A |
| chr3_+_119597874 | 1.25 |
ENST00000488919.5
ENST00000273371.9 ENST00000495992.5 |
PLA1A
|
phospholipase A1 member A |
| chr12_+_48106094 | 1.25 |
ENST00000546755.5
ENST00000549366.5 ENST00000642730.1 ENST00000552792.5 |
PFKM
|
phosphofructokinase, muscle |
| chr22_+_22327298 | 1.25 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr14_-_105987068 | 1.23 |
ENST00000390594.3
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr4_+_94455245 | 1.21 |
ENST00000508216.5
ENST00000514743.5 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr20_+_45406035 | 1.19 |
ENST00000372723.7
ENST00000372722.7 |
DBNDD2
|
dysbindin domain containing 2 |
| chr2_+_89851723 | 1.19 |
ENST00000429992.2
|
IGKV2D-40
|
immunoglobulin kappa variable 2D-40 |
| chr14_-_106771020 | 1.18 |
ENST00000617374.2
|
IGHV2-70
|
immunoglobulin heavy variable 2-70 |
| chr22_+_22322452 | 1.17 |
ENST00000390290.3
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr10_-_60139366 | 1.15 |
ENST00000373815.5
|
ANK3
|
ankyrin 3 |
| chr6_-_41734160 | 1.14 |
ENST00000424495.2
ENST00000420312.6 |
TFEB
|
transcription factor EB |
| chr8_+_2045058 | 1.14 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr22_+_22818994 | 1.13 |
ENST00000390316.2
|
IGLV3-9
|
immunoglobulin lambda variable 3-9 |
| chr22_+_22880706 | 1.13 |
ENST00000390319.2
|
IGLV3-1
|
immunoglobulin lambda variable 3-1 |
| chr6_+_122779707 | 1.13 |
ENST00000368444.8
ENST00000356535.4 |
FABP7
|
fatty acid binding protein 7 |
| chr19_-_55179390 | 1.11 |
ENST00000590851.5
|
SYT5
|
synaptotagmin 5 |
| chr2_-_218831791 | 1.10 |
ENST00000439262.6
ENST00000430489.1 |
PRKAG3
|
protein kinase AMP-activated non-catalytic subunit gamma 3 |
| chr12_-_110920710 | 1.09 |
ENST00000546404.1
|
MYL2
|
myosin light chain 2 |
| chr14_-_105626066 | 1.09 |
ENST00000641978.1
ENST00000390543.3 |
IGHG4
|
immunoglobulin heavy constant gamma 4 (G4m marker) |
| chr20_+_45408276 | 1.08 |
ENST00000372710.5
ENST00000443296.1 |
DBNDD2
|
dysbindin domain containing 2 |
| chr16_+_76277269 | 1.08 |
ENST00000377504.8
ENST00000307431.12 |
CNTNAP4
|
contactin associated protein family member 4 |
| chrX_+_10158448 | 1.07 |
ENST00000380829.5
ENST00000421085.7 ENST00000674669.1 ENST00000454850.1 |
CLCN4
|
chloride voltage-gated channel 4 |
| chr3_-_12158901 | 1.06 |
ENST00000287814.5
|
TIMP4
|
TIMP metallopeptidase inhibitor 4 |
| chr14_-_21022258 | 1.04 |
ENST00000556366.5
|
NDRG2
|
NDRG family member 2 |
| chr1_+_159204860 | 1.03 |
ENST00000368122.4
ENST00000368121.6 |
ACKR1
|
atypical chemokine receptor 1 (Duffy blood group) |
| chr19_-_42232645 | 1.03 |
ENST00000678040.1
ENST00000678524.1 |
GSK3A
|
glycogen synthase kinase 3 alpha |
| chr2_+_90114838 | 1.02 |
ENST00000417279.3
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 |
| chr12_-_21334858 | 1.02 |
ENST00000445053.1
ENST00000458504.5 ENST00000422327.5 ENST00000683939.1 |
SLCO1A2
|
solute carrier organic anion transporter family member 1A2 |
| chr9_-_34710069 | 1.01 |
ENST00000378792.1
ENST00000259607.7 |
CCL21
|
C-C motif chemokine ligand 21 |
| chr19_-_50968775 | 1.01 |
ENST00000391808.5
|
KLK6
|
kallikrein related peptidase 6 |
| chr14_-_106675544 | 1.01 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr5_-_176630364 | 1.00 |
ENST00000310112.7
|
SNCB
|
synuclein beta |
| chr2_-_88966767 | 1.00 |
ENST00000464162.1
|
IGKV1-6
|
immunoglobulin kappa variable 1-6 |
| chr11_-_94401392 | 1.00 |
ENST00000539203.2
ENST00000243673.7 |
GPR83
|
G protein-coupled receptor 83 |
| chr14_-_74923234 | 0.99 |
ENST00000556776.1
ENST00000557413.6 ENST00000555647.5 |
RPS6KL1
|
ribosomal protein S6 kinase like 1 |
| chr9_-_89405926 | 0.98 |
ENST00000433650.5
|
SEMA4D
|
semaphorin 4D |
| chr14_-_106277039 | 0.98 |
ENST00000390610.2
|
IGHV1-24
|
immunoglobulin heavy variable 1-24 |
| chr10_-_118595637 | 0.97 |
ENST00000239032.4
|
PRLHR
|
prolactin releasing hormone receptor |
| chr19_+_49877694 | 0.97 |
ENST00000221543.10
|
TBC1D17
|
TBC1 domain family member 17 |
| chr19_+_49878052 | 0.95 |
ENST00000599049.6
|
TBC1D17
|
TBC1 domain family member 17 |
| chr1_-_100178215 | 0.93 |
ENST00000370138.1
ENST00000370137.6 ENST00000342895.7 ENST00000620882.4 |
LRRC39
|
leucine rich repeat containing 39 |
| chr11_-_3671367 | 0.91 |
ENST00000534359.1
ENST00000250699.2 |
CHRNA10
|
cholinergic receptor nicotinic alpha 10 subunit |
| chr14_-_106422175 | 0.91 |
ENST00000390619.2
|
IGHV4-39
|
immunoglobulin heavy variable 4-39 |
| chr14_+_76376113 | 0.91 |
ENST00000644823.1
|
ESRRB
|
estrogen related receptor beta |
| chr10_-_95422012 | 0.91 |
ENST00000486141.3
|
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr1_+_222739670 | 0.90 |
ENST00000456298.5
|
FAM177B
|
family with sequence similarity 177 member B |
| chr11_+_109422174 | 0.89 |
ENST00000327419.7
|
C11orf87
|
chromosome 11 open reading frame 87 |
| chr11_+_1839602 | 0.88 |
ENST00000617947.4
ENST00000252898.11 |
TNNI2
|
troponin I2, fast skeletal type |
| chr11_+_1839452 | 0.88 |
ENST00000381906.5
|
TNNI2
|
troponin I2, fast skeletal type |
| chr14_-_106737547 | 0.87 |
ENST00000632209.1
|
IGHV1-69-2
|
immunoglobulin heavy variable 1-69-2 |
| chr3_+_69936629 | 0.87 |
ENST00000394348.2
ENST00000531774.1 |
MITF
|
melanocyte inducing transcription factor |
| chr11_+_67404077 | 0.86 |
ENST00000542590.2
ENST00000312390.9 |
TBC1D10C
|
TBC1 domain family member 10C |
| chr5_-_142686079 | 0.85 |
ENST00000337706.7
|
FGF1
|
fibroblast growth factor 1 |
| chr14_-_106875069 | 0.85 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr8_-_90082871 | 0.82 |
ENST00000265431.7
|
CALB1
|
calbindin 1 |
| chr11_-_111449981 | 0.82 |
ENST00000531398.1
|
POU2AF1
|
POU class 2 homeobox associating factor 1 |
| chr14_-_21022095 | 0.82 |
ENST00000635386.1
|
NDRG2
|
NDRG family member 2 |
| chr1_-_32870775 | 0.81 |
ENST00000649537.2
ENST00000373471.9 |
FNDC5
|
fibronectin type III domain containing 5 |
| chr2_+_26970628 | 0.80 |
ENST00000233121.7
ENST00000405074.7 |
MAPRE3
|
microtubule associated protein RP/EB family member 3 |
| chr12_-_122395422 | 0.80 |
ENST00000540304.6
|
CLIP1
|
CAP-Gly domain containing linker protein 1 |
| chr18_-_3219849 | 0.80 |
ENST00000261606.11
|
MYOM1
|
myomesin 1 |
| chr6_-_41705813 | 0.80 |
ENST00000419574.6
ENST00000445214.2 |
TFEB
|
transcription factor EB |
| chr16_-_58734299 | 0.80 |
ENST00000245206.10
ENST00000434819.2 |
GOT2
|
glutamic-oxaloacetic transaminase 2 |
| chr11_+_67403887 | 0.79 |
ENST00000526387.5
|
TBC1D10C
|
TBC1 domain family member 10C |
| chr18_-_3219961 | 0.79 |
ENST00000356443.9
|
MYOM1
|
myomesin 1 |
| chr11_-_118342645 | 0.78 |
ENST00000529594.5
|
CD3D
|
CD3d molecule |
| chr14_-_34714538 | 0.78 |
ENST00000672163.1
|
CFL2
|
cofilin 2 |
| chr8_-_109975757 | 0.77 |
ENST00000524391.6
|
KCNV1
|
potassium voltage-gated channel modifier subfamily V member 1 |
| chr16_+_66366675 | 0.76 |
ENST00000341529.8
ENST00000649567.1 |
CDH5
|
cadherin 5 |
| chr2_-_157325659 | 0.76 |
ENST00000409925.1
|
ERMN
|
ermin |
| chr8_+_119873710 | 0.75 |
ENST00000523492.5
ENST00000286234.6 |
DEPTOR
|
DEP domain containing MTOR interacting protein |
| chr22_+_22588155 | 0.75 |
ENST00000390302.3
|
IGLV2-33
|
immunoglobulin lambda variable 2-33 (non-functional) |
| chr22_+_22380766 | 0.75 |
ENST00000390297.3
|
IGLV1-44
|
immunoglobulin lambda variable 1-44 |
| chr7_+_142349135 | 0.75 |
ENST00000634383.1
|
TRBV6-2
|
T cell receptor beta variable 6-2 |
| chr14_-_21022432 | 0.75 |
ENST00000557633.5
|
NDRG2
|
NDRG family member 2 |
| chr10_-_20897288 | 0.74 |
ENST00000377122.9
|
NEBL
|
nebulette |
| chr22_+_21015027 | 0.74 |
ENST00000413302.6
ENST00000401443.5 |
P2RX6
|
purinergic receptor P2X 6 |
| chr14_+_91114026 | 0.74 |
ENST00000521081.5
ENST00000520328.5 ENST00000524232.5 ENST00000522170.5 ENST00000256324.15 ENST00000519950.5 ENST00000523879.5 ENST00000521077.6 ENST00000518665.6 |
DGLUCY
|
D-glutamate cyclase |
| chr3_+_69936583 | 0.74 |
ENST00000314557.10
ENST00000394351.9 |
MITF
|
melanocyte inducing transcription factor |
| chr11_+_62270150 | 0.74 |
ENST00000525380.1
ENST00000227918.3 |
SCGB2A2
|
secretoglobin family 2A member 2 |
| chr18_+_9103959 | 0.73 |
ENST00000400033.1
|
NDUFV2
|
NADH:ubiquinone oxidoreductase core subunit V2 |
| chr17_+_7438267 | 0.73 |
ENST00000575235.5
|
FGF11
|
fibroblast growth factor 11 |
| chrX_-_153794494 | 0.72 |
ENST00000370092.7
ENST00000619865.4 |
IDH3G
|
isocitrate dehydrogenase (NAD(+)) 3 non-catalytic subunit gamma |
| chr4_-_99657820 | 0.72 |
ENST00000511828.2
|
C4orf54
|
chromosome 4 open reading frame 54 |
| chr6_-_46954922 | 0.72 |
ENST00000265417.7
|
ADGRF5
|
adhesion G protein-coupled receptor F5 |
| chr16_+_76277393 | 0.72 |
ENST00000611870.5
|
CNTNAP4
|
contactin associated protein family member 4 |
| chr3_+_173397725 | 0.71 |
ENST00000413821.1
|
NLGN1
|
neuroligin 1 |
| chr17_-_75855204 | 0.71 |
ENST00000589642.5
ENST00000593002.1 ENST00000590221.5 ENST00000587374.5 ENST00000585462.5 ENST00000254806.8 ENST00000433525.6 ENST00000626827.2 |
WBP2
|
WW domain binding protein 2 |
| chr16_+_32066065 | 0.71 |
ENST00000354689.6
|
IGHV3OR16-9
|
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr3_+_167735238 | 0.71 |
ENST00000472941.5
|
SERPINI1
|
serpin family I member 1 |
| chr11_+_46345368 | 0.71 |
ENST00000528615.5
|
DGKZ
|
diacylglycerol kinase zeta |
| chrX_-_153794356 | 0.70 |
ENST00000427365.6
ENST00000444450.5 ENST00000217901.10 ENST00000370093.5 |
IDH3G
|
isocitrate dehydrogenase (NAD(+)) 3 non-catalytic subunit gamma |
| chr17_+_69502397 | 0.70 |
ENST00000613873.4
ENST00000589647.5 |
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr2_-_89143133 | 0.69 |
ENST00000492167.1
|
IGKV3-20
|
immunoglobulin kappa variable 3-20 |
| chr7_+_142345412 | 0.69 |
ENST00000390392.3
|
TRBV4-2
|
T cell receptor beta variable 4-2 |
| chr2_+_154697855 | 0.69 |
ENST00000651198.1
|
KCNJ3
|
potassium inwardly rectifying channel subfamily J member 3 |
| chr21_-_10649835 | 0.69 |
ENST00000622028.1
|
IGHV1OR21-1
|
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr22_+_22822658 | 0.68 |
ENST00000620395.2
|
IGLV2-8
|
immunoglobulin lambda variable 2-8 |
| chr14_+_104581141 | 0.68 |
ENST00000410013.1
|
C14orf180
|
chromosome 14 open reading frame 180 |
| chr4_-_113517607 | 0.68 |
ENST00000509594.1
|
CAMK2D
|
calcium/calmodulin dependent protein kinase II delta |
| chr14_-_106211453 | 0.68 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr1_-_157700738 | 0.67 |
ENST00000368186.9
ENST00000496769.1 ENST00000368184.8 |
FCRL3
|
Fc receptor like 3 |
| chr3_+_111998739 | 0.67 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr14_-_34714549 | 0.66 |
ENST00000555765.5
ENST00000672517.1 |
CFL2
|
cofilin 2 |
| chr5_+_176630618 | 0.66 |
ENST00000647833.1
ENST00000318682.11 |
EIF4E1B
|
eukaryotic translation initiation factor 4E family member 1B |
| chr12_+_109131350 | 0.66 |
ENST00000539864.1
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr14_+_101964561 | 0.66 |
ENST00000643508.2
ENST00000680137.1 ENST00000644881.2 ENST00000645149.2 ENST00000681574.1 ENST00000360184.10 ENST00000679720.1 ENST00000645114.2 |
DYNC1H1
|
dynein cytoplasmic 1 heavy chain 1 |
| chr13_-_80341100 | 0.65 |
ENST00000377104.4
|
SPRY2
|
sprouty RTK signaling antagonist 2 |
| chr3_-_112499457 | 0.65 |
ENST00000334529.10
|
BTLA
|
B and T lymphocyte associated |
| chr19_-_6502330 | 0.65 |
ENST00000598006.1
ENST00000601152.5 |
TUBB4A
|
tubulin beta 4A class IVa |
| chr8_+_22059198 | 0.65 |
ENST00000523266.5
ENST00000519907.5 |
DMTN
|
dematin actin binding protein |
| chr22_+_22792485 | 0.65 |
ENST00000390314.2
|
IGLV2-11
|
immunoglobulin lambda variable 2-11 |
| chr2_-_197676012 | 0.65 |
ENST00000429081.1
|
RFTN2
|
raftlin family member 2 |
| chr19_-_6502301 | 0.65 |
ENST00000264071.7
ENST00000594276.5 ENST00000594075.5 ENST00000600216.5 ENST00000596926.5 |
TUBB4A
|
tubulin beta 4A class IVa |
| chr5_+_10564064 | 0.64 |
ENST00000296657.7
|
ANKRD33B
|
ankyrin repeat domain 33B |
| chr19_-_45902594 | 0.64 |
ENST00000322217.6
|
MYPOP
|
Myb related transcription factor, partner of profilin |
| chr22_+_22899481 | 0.64 |
ENST00000390322.2
|
IGLJ2
|
immunoglobulin lambda joining 2 |
| chr19_-_6501766 | 0.64 |
ENST00000596291.1
|
TUBB4A
|
tubulin beta 4A class IVa |
| chr19_-_50968966 | 0.64 |
ENST00000376851.7
|
KLK6
|
kallikrein related peptidase 6 |
| chr14_-_34714579 | 0.64 |
ENST00000298159.11
|
CFL2
|
cofilin 2 |
| chr12_+_12785652 | 0.63 |
ENST00000356591.5
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr11_+_65576023 | 0.62 |
ENST00000533237.5
ENST00000309295.9 ENST00000634639.1 |
EHBP1L1
|
EH domain binding protein 1 like 1 |
| chr1_+_202462730 | 0.62 |
ENST00000290419.9
ENST00000491336.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr12_-_54984667 | 0.62 |
ENST00000524668.5
ENST00000533607.1 ENST00000449076.6 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr6_-_89315291 | 0.61 |
ENST00000402938.4
|
GABRR2
|
gamma-aminobutyric acid type A receptor subunit rho2 |
| chr1_-_217137748 | 0.61 |
ENST00000493603.5
ENST00000366940.6 |
ESRRG
|
estrogen related receptor gamma |
| chr6_-_41736239 | 0.61 |
ENST00000358871.6
|
TFEB
|
transcription factor EB |
| chr2_+_115162107 | 0.61 |
ENST00000310323.12
|
DPP10
|
dipeptidyl peptidase like 10 |
| chr14_+_63204859 | 0.61 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J |
| chr14_-_34713759 | 0.60 |
ENST00000673315.1
|
CFL2
|
cofilin 2 |
| chr15_-_72271244 | 0.60 |
ENST00000287196.13
|
PARP6
|
poly(ADP-ribose) polymerase family member 6 |
| chr6_+_37005630 | 0.60 |
ENST00000274963.13
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr3_-_170586056 | 0.59 |
ENST00000231706.6
|
SLC7A14
|
solute carrier family 7 member 14 |
| chr19_-_4722767 | 0.59 |
ENST00000600621.5
|
DPP9
|
dipeptidyl peptidase 9 |
| chr19_-_49878350 | 0.59 |
ENST00000391834.6
ENST00000391830.1 |
AKT1S1
|
AKT1 substrate 1 |
| chr19_+_38390055 | 0.59 |
ENST00000587947.5
ENST00000338502.8 |
SPRED3
|
sprouty related EVH1 domain containing 3 |
| chr14_-_34713788 | 0.59 |
ENST00000341223.8
|
CFL2
|
cofilin 2 |
| chr9_+_93289187 | 0.58 |
ENST00000453718.2
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr17_+_69502255 | 0.58 |
ENST00000588110.5
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr16_+_58501468 | 0.58 |
ENST00000566656.5
ENST00000566618.5 |
NDRG4
|
NDRG family member 4 |
| chr3_-_112499358 | 0.58 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr15_-_89751292 | 0.57 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior bHLH transcription factor 1 |
| chr14_+_91114431 | 0.57 |
ENST00000428926.6
ENST00000517362.5 |
DGLUCY
|
D-glutamate cyclase |
| chr17_+_44846318 | 0.57 |
ENST00000591513.5
|
HIGD1B
|
HIG1 hypoxia inducible domain family member 1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of CUX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 1.1 | 3.3 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 1.0 | 9.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.7 | 2.2 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.6 | 3.7 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.5 | 1.6 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.5 | 1.4 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.5 | 1.8 | GO:1904980 | positive regulation of endosome organization(GO:1904980) |
| 0.4 | 1.3 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.4 | 1.3 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.4 | 2.6 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.4 | 2.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.4 | 1.8 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.3 | 2.8 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.3 | 1.0 | GO:0010905 | regulation of UDP-glucose catabolic process(GO:0010904) negative regulation of UDP-glucose catabolic process(GO:0010905) regulation of glycogen synthase activity, transferring glucose-1-phosphate(GO:1904226) negative regulation of glycogen synthase activity, transferring glucose-1-phosphate(GO:1904227) |
| 0.3 | 2.7 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.3 | 3.7 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.3 | 0.8 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.3 | 0.8 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.3 | 0.8 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.3 | 2.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 0.9 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.2 | 1.3 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.2 | 1.3 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.2 | 1.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 1.5 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.2 | 3.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 0.6 | GO:0090381 | cardiac cell fate determination(GO:0060913) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.2 | 1.3 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 0.6 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 0.2 | 1.5 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 2.1 | GO:0060125 | habituation(GO:0046959) negative regulation of growth hormone secretion(GO:0060125) |
| 0.2 | 0.7 | GO:0099545 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.2 | 1.0 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.2 | 0.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.2 | 0.5 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.2 | 1.4 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 1.7 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.2 | 0.5 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.2 | 0.5 | GO:1902616 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.2 | 1.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.3 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.1 | 0.4 | GO:2000984 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.1 | 0.4 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.1 | 0.6 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.3 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 1.5 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 2.6 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.8 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.6 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 1.3 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 1.7 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 1.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 2.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 1.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.7 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.1 | 1.4 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.1 | 0.3 | GO:1902309 | regulation of heart rate by hormone(GO:0003064) negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 1.2 | GO:0098907 | protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.1 | 17.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.6 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 1.1 | GO:1901731 | positive regulation of platelet aggregation(GO:1901731) |
| 0.1 | 0.7 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.4 | GO:0061582 | colon epithelial cell migration(GO:0061580) intestinal epithelial cell migration(GO:0061582) |
| 0.1 | 0.4 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 0.7 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.6 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 0.5 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.8 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 0.5 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.1 | 0.5 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.1 | 0.9 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.4 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.1 | 0.7 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.3 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.1 | 1.9 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.1 | 0.7 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.3 | GO:0002290 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.1 | 2.3 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 0.4 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.1 | 0.3 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 1.3 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.1 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 1.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.8 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.4 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.5 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.1 | 0.3 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.1 | 0.3 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.1 | 1.1 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 0.9 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.3 | GO:0031106 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.1 | 1.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.7 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.1 | 0.8 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 0.8 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 1.3 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 1.8 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 0.7 | GO:0043129 | surfactant homeostasis(GO:0043129) positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.1 | 0.5 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.3 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 0.4 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 0.3 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 0.2 | GO:0014028 | aortic valve development(GO:0003176) aortic valve morphogenesis(GO:0003180) notochord formation(GO:0014028) |
| 0.1 | 0.3 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 0.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) type B pancreatic cell maturation(GO:0072560) |
| 0.1 | 0.7 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.3 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 0.8 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 1.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.7 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.3 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.1 | 1.0 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.2 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.1 | 0.5 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.5 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 4.2 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 0.5 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.5 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 2.2 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.5 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.5 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.5 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.3 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.6 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.3 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 0.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.3 | GO:0072011 | glomerular endothelium development(GO:0072011) |
| 0.0 | 0.4 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.4 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.4 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 1.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.2 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.2 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.5 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 2.2 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.0 | 0.3 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) |
| 0.0 | 0.3 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.2 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.4 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.2 | GO:0035912 | dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.6 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.5 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 7.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.7 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.5 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.7 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.7 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.2 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 1.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.4 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 1.8 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.5 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.9 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.4 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.4 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.0 | GO:1903937 | response to acrylamide(GO:1903937) |
| 0.0 | 0.2 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 0.3 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 1.6 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.4 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.0 | 0.4 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 1.5 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.6 | GO:0060973 | cell migration involved in heart development(GO:0060973) |
| 0.0 | 0.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.4 | GO:0046641 | positive regulation of alpha-beta T cell proliferation(GO:0046641) |
| 0.0 | 0.1 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.0 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0072708 | response to sorbitol(GO:0072708) |
| 0.0 | 2.1 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0043380 | memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) |
| 0.0 | 1.3 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.1 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 | 0.1 | GO:0006169 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
| 0.0 | 0.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 1.9 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 1.1 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.7 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.9 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 1.1 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.5 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.4 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.0 | GO:0035977 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.4 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.0 | 1.1 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.5 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 2.7 | GO:0050709 | negative regulation of protein secretion(GO:0050709) |
| 0.0 | 0.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.3 | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.0 | 1.7 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 2.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 1.7 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.8 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.2 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 1.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.0 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.0 | GO:0032499 | detection of peptidoglycan(GO:0032499) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.8 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.9 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.2 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) chromatin maintenance(GO:0070827) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.3 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.0 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 0.0 | 0.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.3 | GO:0042092 | type 2 immune response(GO:0042092) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.9 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 1.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.2 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.3 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 0.5 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 2.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.2 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 1.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.3 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.8 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.8 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.2 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.0 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.6 | 11.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.5 | 2.3 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.4 | 1.6 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.4 | 4.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.4 | 2.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.3 | 4.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.3 | 3.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 1.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.2 | 1.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.2 | 0.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 0.9 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 1.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 9.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 0.5 | GO:0090651 | apical cytoplasm(GO:0090651) |
| 0.2 | 1.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.7 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 3.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 1.0 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 1.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 1.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.3 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.1 | 1.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 9.9 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 2.1 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 0.8 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.2 | GO:1990696 | stereocilia ankle link(GO:0002141) USH2 complex(GO:1990696) |
| 0.1 | 0.5 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.1 | 0.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.3 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.1 | 1.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 1.4 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.2 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.1 | 0.9 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.8 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 1.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 1.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 1.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 2.0 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 3.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.5 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.8 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 1.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 2.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.8 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 1.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 4.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.8 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 2.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 3.5 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 1.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 2.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.6 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.4 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 2.9 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 1.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 11.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.4 | 3.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.4 | 1.3 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.4 | 2.4 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.4 | 1.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.4 | 1.4 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.3 | 0.8 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.2 | 1.8 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 1.3 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 1.9 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.2 | 1.8 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 2.0 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.2 | 0.6 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.2 | 3.3 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 1.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 0.5 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.2 | 1.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 1.5 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.2 | 1.0 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.2 | 1.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 0.5 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.2 | 3.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 0.5 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.2 | 2.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 1.3 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 0.5 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.8 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 1.3 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.5 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 8.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.4 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.1 | 1.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.8 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.9 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 3.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.7 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.5 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.1 | 2.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.4 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.4 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.1 | 2.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.3 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.1 | 0.4 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.1 | 0.4 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 0.4 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.6 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.4 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.6 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 1.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.5 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.3 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.3 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 0.2 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.1 | 0.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 2.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.6 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 12.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 2.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.2 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 1.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.2 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 0.2 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.1 | 1.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.2 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 4.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.3 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 0.5 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.2 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.4 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 1.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.6 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 3.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 6.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 1.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.9 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.4 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 2.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 1.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.5 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 1.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.2 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.8 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.7 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.0 | 1.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.2 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 4.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 2.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 1.6 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 0.8 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 1.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 2.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 2.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 2.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 2.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 17.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.9 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 1.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.8 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 1.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 1.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 1.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.7 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 1.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 2.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.5 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.0 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 1.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.0 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 2.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |