Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for DDIT3
Z-value: 0.85
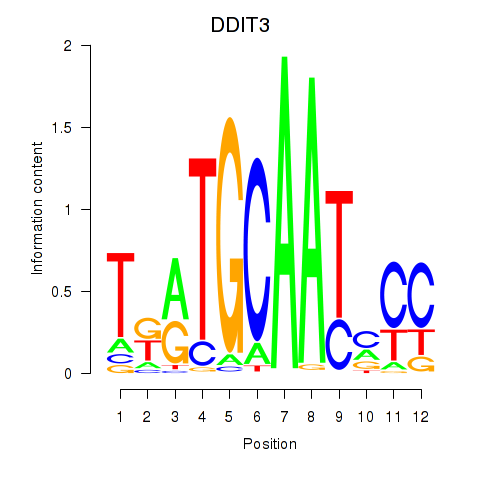
Transcription factors associated with DDIT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DDIT3
|
ENSG00000175197.13 | DNA damage inducible transcript 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DDIT3 | hg38_v1_chr12_-_57520480_57520521 | -0.30 | 9.5e-02 | Click! |
Activity profile of DDIT3 motif
Sorted Z-values of DDIT3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_90921079 | 7.30 |
ENST00000371697.4
|
ANKRD1
|
ankyrin repeat domain 1 |
| chr2_-_178807415 | 5.04 |
ENST00000342992.10
ENST00000460472.6 ENST00000589042.5 ENST00000591111.5 ENST00000360870.10 |
TTN
|
titin |
| chr10_-_114685000 | 2.56 |
ENST00000369256.6
|
ABLIM1
|
actin binding LIM protein 1 |
| chr10_-_114684612 | 2.41 |
ENST00000533213.6
ENST00000369252.8 |
ABLIM1
|
actin binding LIM protein 1 |
| chr10_-_114684457 | 2.29 |
ENST00000392955.7
|
ABLIM1
|
actin binding LIM protein 1 |
| chr17_+_4951758 | 2.10 |
ENST00000518175.1
|
ENO3
|
enolase 3 |
| chr15_+_85380672 | 1.97 |
ENST00000361243.6
ENST00000560256.1 |
AKAP13
|
A-kinase anchoring protein 13 |
| chr15_+_85380565 | 1.83 |
ENST00000559362.5
ENST00000394518.7 |
AKAP13
|
A-kinase anchoring protein 13 |
| chr14_+_32329341 | 1.68 |
ENST00000557354.5
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A-kinase anchoring protein 6 |
| chr5_-_178590367 | 1.59 |
ENST00000390654.8
|
COL23A1
|
collagen type XXIII alpha 1 chain |
| chr14_+_32329256 | 1.56 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6 |
| chr10_-_17617326 | 1.56 |
ENST00000326961.6
ENST00000361271.8 |
HACD1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr10_-_17617235 | 1.48 |
ENST00000466335.1
|
HACD1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr2_+_63588992 | 1.41 |
ENST00000454035.5
|
MDH1
|
malate dehydrogenase 1 |
| chr6_+_144285681 | 1.23 |
ENST00000433557.1
|
UTRN
|
utrophin |
| chr22_+_31122923 | 1.23 |
ENST00000620191.4
ENST00000412277.6 ENST00000412985.5 ENST00000331075.10 ENST00000420017.5 ENST00000400294.6 ENST00000405300.5 ENST00000404390.7 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr2_+_63588953 | 1.14 |
ENST00000409908.5
ENST00000442225.5 ENST00000233114.13 ENST00000539945.7 ENST00000409476.5 ENST00000436321.5 |
MDH1
|
malate dehydrogenase 1 |
| chr2_+_63589135 | 1.13 |
ENST00000432309.6
|
MDH1
|
malate dehydrogenase 1 |
| chr7_+_80626148 | 1.09 |
ENST00000428497.5
|
CD36
|
CD36 molecule |
| chr15_-_63833911 | 1.08 |
ENST00000560462.1
ENST00000558532.1 ENST00000561400.1 ENST00000443617.7 |
HERC1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr6_+_12716760 | 0.95 |
ENST00000332995.12
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr15_+_85380625 | 0.94 |
ENST00000560302.5
|
AKAP13
|
A-kinase anchoring protein 13 |
| chr2_-_40453438 | 0.93 |
ENST00000455476.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr6_+_12716554 | 0.92 |
ENST00000676159.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr12_+_32485963 | 0.85 |
ENST00000531134.7
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr1_+_174875505 | 0.85 |
ENST00000486220.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr2_-_63588725 | 0.64 |
ENST00000431065.1
|
WDPCP
|
WD repeat containing planar cell polarity effector |
| chr19_+_48755512 | 0.64 |
ENST00000593756.6
|
FGF21
|
fibroblast growth factor 21 |
| chr14_+_101809795 | 0.63 |
ENST00000350249.7
ENST00000557621.5 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2 regulatory subunit B'gamma |
| chr13_+_32031706 | 0.59 |
ENST00000542859.6
|
FRY
|
FRY microtubule binding protein |
| chr14_+_101809855 | 0.56 |
ENST00000557714.1
ENST00000445439.7 ENST00000334743.9 ENST00000557095.5 |
PPP2R5C
|
protein phosphatase 2 regulatory subunit B'gamma |
| chr6_+_12716801 | 0.56 |
ENST00000674595.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr8_-_18887018 | 0.55 |
ENST00000523619.5
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr5_+_112738331 | 0.53 |
ENST00000512211.6
|
APC
|
APC regulator of WNT signaling pathway |
| chr1_+_89843421 | 0.53 |
ENST00000527156.1
|
LRRC8D
|
leucine rich repeat containing 8 VRAC subunit D |
| chr11_+_18602969 | 0.53 |
ENST00000636011.1
ENST00000542172.1 |
SPTY2D1OS
|
SPTY2D1 opposite strand |
| chr18_-_57621741 | 0.50 |
ENST00000587194.1
ENST00000591599.5 ENST00000588661.5 ENST00000256854.10 |
NARS1
|
asparaginyl-tRNA synthetase 1 |
| chr1_+_61203496 | 0.48 |
ENST00000663597.1
|
NFIA
|
nuclear factor I A |
| chr17_+_58238694 | 0.47 |
ENST00000581008.1
|
LPO
|
lactoperoxidase |
| chr20_+_56412112 | 0.46 |
ENST00000360314.7
|
CASS4
|
Cas scaffold protein family member 4 |
| chr14_+_21797272 | 0.44 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1 |
| chr5_+_146203544 | 0.42 |
ENST00000506502.2
|
ENSG00000275740.1
|
novel readthrough transcript |
| chr1_-_149861210 | 0.42 |
ENST00000579512.2
|
H4C15
|
H4 clustered histone 15 |
| chr21_-_30492008 | 0.41 |
ENST00000334063.6
|
KRTAP19-3
|
keratin associated protein 19-3 |
| chr20_+_56412393 | 0.40 |
ENST00000679529.1
|
CASS4
|
Cas scaffold protein family member 4 |
| chr3_-_122993232 | 0.40 |
ENST00000650207.1
ENST00000616742.4 ENST00000393583.6 |
SEMA5B
|
semaphorin 5B |
| chr9_-_92293674 | 0.40 |
ENST00000683679.1
ENST00000683565.1 ENST00000684557.1 ENST00000682578.1 ENST00000443024.7 ENST00000375643.7 ENST00000683469.1 |
IARS1
|
isoleucyl-tRNA synthetase 1 |
| chr12_-_7747339 | 0.38 |
ENST00000543765.1
ENST00000360345.8 ENST00000540085.5 |
CLEC4C
|
C-type lectin domain family 4 member C |
| chr7_-_44490609 | 0.38 |
ENST00000355451.8
|
NUDCD3
|
NudC domain containing 3 |
| chr8_-_21812320 | 0.37 |
ENST00000517328.5
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr5_+_72848161 | 0.37 |
ENST00000506351.6
|
TNPO1
|
transportin 1 |
| chr3_+_1093002 | 0.37 |
ENST00000446702.7
|
CNTN6
|
contactin 6 |
| chr12_-_52680398 | 0.35 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr3_-_56468346 | 0.35 |
ENST00000288221.11
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr19_-_6459735 | 0.35 |
ENST00000334510.9
ENST00000301454.9 |
SLC25A23
|
solute carrier family 25 member 23 |
| chr14_-_22976812 | 0.32 |
ENST00000553592.5
|
AJUBA
|
ajuba LIM protein |
| chr1_+_113905156 | 0.31 |
ENST00000650596.1
|
DCLRE1B
|
DNA cross-link repair 1B |
| chr9_+_109780312 | 0.31 |
ENST00000483909.5
ENST00000413420.5 ENST00000302798.7 |
PALM2AKAP2
|
PALM2 and AKAP2 fusion |
| chr17_-_49848017 | 0.29 |
ENST00000326219.5
ENST00000334568.8 ENST00000352793.6 ENST00000398154.5 ENST00000436235.5 |
TAC4
|
tachykinin precursor 4 |
| chr1_-_152224193 | 0.29 |
ENST00000368801.4
|
HRNR
|
hornerin |
| chr1_+_111227672 | 0.29 |
ENST00000474304.6
|
CHI3L2
|
chitinase 3 like 2 |
| chr2_-_63588390 | 0.28 |
ENST00000272321.12
ENST00000409562.7 |
WDPCP
|
WD repeat containing planar cell polarity effector |
| chr1_+_149832647 | 0.27 |
ENST00000578186.2
|
H4C14
|
H4 clustered histone 14 |
| chr4_+_183659267 | 0.27 |
ENST00000357207.8
ENST00000334690.11 |
TRAPPC11
|
trafficking protein particle complex 11 |
| chr14_-_22976793 | 0.27 |
ENST00000553911.1
|
AJUBA
|
ajuba LIM protein |
| chr18_-_63158208 | 0.26 |
ENST00000678301.1
|
BCL2
|
BCL2 apoptosis regulator |
| chr21_-_44914271 | 0.26 |
ENST00000522931.5
|
ITGB2
|
integrin subunit beta 2 |
| chr10_-_102114935 | 0.24 |
ENST00000361198.9
|
LDB1
|
LIM domain binding 1 |
| chr10_-_24706622 | 0.22 |
ENST00000680286.1
|
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr5_+_119476530 | 0.22 |
ENST00000645099.1
ENST00000513628.5 |
HSD17B4
|
hydroxysteroid 17-beta dehydrogenase 4 |
| chr11_-_10294194 | 0.22 |
ENST00000676387.1
ENST00000256190.13 ENST00000675281.1 |
SBF2
|
SET binding factor 2 |
| chr1_-_19923617 | 0.22 |
ENST00000375116.3
|
PLA2G2E
|
phospholipase A2 group IIE |
| chr4_-_98657635 | 0.21 |
ENST00000515287.5
ENST00000511651.5 ENST00000505184.5 |
TSPAN5
|
tetraspanin 5 |
| chrX_-_52517213 | 0.21 |
ENST00000375616.5
|
XAGE1B
|
X antigen family member 1B |
| chrX_+_52495791 | 0.21 |
ENST00000375602.2
ENST00000375600.5 |
XAGE1A
|
X antigen family member 1A |
| chr15_+_40952962 | 0.20 |
ENST00000444189.7
|
CHAC1
|
ChaC glutathione specific gamma-glutamylcyclotransferase 1 |
| chr2_+_226835936 | 0.19 |
ENST00000341329.7
ENST00000437454.5 ENST00000443477.5 ENST00000423616.1 ENST00000392062.7 ENST00000448992.5 |
RHBDD1
|
rhomboid domain containing 1 |
| chr7_+_44000889 | 0.18 |
ENST00000258704.3
|
SPDYE1
|
speedy/RINGO cell cycle regulator family member E1 |
| chr5_-_132295236 | 0.18 |
ENST00000431054.5
|
P4HA2
|
prolyl 4-hydroxylase subunit alpha 2 |
| chr6_-_157323498 | 0.18 |
ENST00000400788.9
ENST00000367144.4 |
TMEM242
|
transmembrane protein 242 |
| chr19_+_1450113 | 0.18 |
ENST00000590469.6
ENST00000590877.5 ENST00000233607.6 |
APC2
|
APC regulator of WNT signaling pathway 2 |
| chr7_-_99408548 | 0.17 |
ENST00000626285.1
ENST00000350498.8 |
PDAP1
|
PDGFA associated protein 1 |
| chr3_-_9553796 | 0.15 |
ENST00000287585.8
|
LHFPL4
|
LHFPL tetraspan subfamily member 4 |
| chr9_+_122723990 | 0.15 |
ENST00000259466.1
|
OR1L4
|
olfactory receptor family 1 subfamily L member 4 |
| chr19_-_35501878 | 0.15 |
ENST00000593342.5
ENST00000601650.1 ENST00000408915.6 |
DMKN
|
dermokine |
| chr2_+_6978624 | 0.15 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr3_+_37975773 | 0.15 |
ENST00000436654.1
|
CTDSPL
|
CTD small phosphatase like |
| chr12_+_64405046 | 0.15 |
ENST00000540203.5
|
XPOT
|
exportin for tRNA |
| chr20_+_56412249 | 0.15 |
ENST00000679887.1
ENST00000434344.2 |
CASS4
|
Cas scaffold protein family member 4 |
| chr13_+_24270841 | 0.14 |
ENST00000454083.1
|
SPATA13
|
spermatogenesis associated 13 |
| chr9_+_107283137 | 0.14 |
ENST00000419616.5
|
RAD23B
|
RAD23 homolog B, nucleotide excision repair protein |
| chr4_+_140257286 | 0.14 |
ENST00000394205.7
|
SCOC
|
short coiled-coil protein |
| chr8_-_92017292 | 0.13 |
ENST00000521553.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr9_+_109780292 | 0.12 |
ENST00000374530.7
|
PALM2AKAP2
|
PALM2 and AKAP2 fusion |
| chr3_+_44799187 | 0.12 |
ENST00000425755.5
|
KIF15
|
kinesin family member 15 |
| chr3_+_9423108 | 0.12 |
ENST00000468208.2
|
SETD5
|
SET domain containing 5 |
| chr12_+_106601137 | 0.11 |
ENST00000357881.8
|
RFX4
|
regulatory factor X4 |
| chr5_+_146203593 | 0.11 |
ENST00000265271.7
|
RBM27
|
RNA binding motif protein 27 |
| chr14_+_21894433 | 0.11 |
ENST00000390438.2
|
TRAV8-4
|
T cell receptor alpha variable 8-4 |
| chr15_-_63833875 | 0.11 |
ENST00000560316.1
|
HERC1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr7_-_56034133 | 0.10 |
ENST00000421626.5
|
PSPH
|
phosphoserine phosphatase |
| chr17_+_58238426 | 0.10 |
ENST00000421678.6
ENST00000262290.9 ENST00000543544.5 |
LPO
|
lactoperoxidase |
| chr9_+_109780179 | 0.10 |
ENST00000314527.9
|
PALM2AKAP2
|
PALM2 and AKAP2 fusion |
| chr1_-_113905020 | 0.10 |
ENST00000432415.5
ENST00000369571.2 ENST00000256658.8 ENST00000369564.5 |
AP4B1
|
adaptor related protein complex 4 subunit beta 1 |
| chr12_-_109309218 | 0.10 |
ENST00000355216.5
|
FOXN4
|
forkhead box N4 |
| chr10_+_100997040 | 0.10 |
ENST00000370223.7
|
LZTS2
|
leucine zipper tumor suppressor 2 |
| chr1_+_111227610 | 0.10 |
ENST00000369744.6
|
CHI3L2
|
chitinase 3 like 2 |
| chr19_+_10625507 | 0.09 |
ENST00000590857.5
ENST00000588688.5 ENST00000586078.5 ENST00000335757.10 |
SLC44A2
|
solute carrier family 44 member 2 |
| chr11_-_69704013 | 0.09 |
ENST00000294312.4
|
FGF19
|
fibroblast growth factor 19 |
| chr1_+_111227699 | 0.09 |
ENST00000369748.9
|
CHI3L2
|
chitinase 3 like 2 |
| chr20_+_6007245 | 0.08 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr17_+_4433904 | 0.08 |
ENST00000355530.7
|
SPNS3
|
sphingolipid transporter 3 (putative) |
| chr11_+_4586791 | 0.08 |
ENST00000641486.1
|
OR52I2
|
olfactory receptor family 52 subfamily I member 2 |
| chr11_+_20022550 | 0.08 |
ENST00000533917.5
|
NAV2
|
neuron navigator 2 |
| chr5_+_154755272 | 0.08 |
ENST00000518297.6
|
LARP1
|
La ribonucleoprotein 1, translational regulator |
| chr7_+_18290089 | 0.08 |
ENST00000433709.6
|
HDAC9
|
histone deacetylase 9 |
| chr2_-_182242031 | 0.07 |
ENST00000358139.6
|
PDE1A
|
phosphodiesterase 1A |
| chr1_+_222737283 | 0.07 |
ENST00000360827.6
|
FAM177B
|
family with sequence similarity 177 member B |
| chrX_+_30235894 | 0.07 |
ENST00000620842.1
|
MAGEB3
|
MAGE family member B3 |
| chr8_+_27771942 | 0.07 |
ENST00000523566.5
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr12_-_121579638 | 0.06 |
ENST00000446152.6
|
KDM2B
|
lysine demethylase 2B |
| chr4_+_157076119 | 0.05 |
ENST00000541722.5
ENST00000264428.9 ENST00000512619.5 |
GLRB
|
glycine receptor beta |
| chr3_+_136957948 | 0.05 |
ENST00000329582.9
|
IL20RB
|
interleukin 20 receptor subunit beta |
| chr1_+_152842851 | 0.05 |
ENST00000431011.3
|
LCE6A
|
late cornified envelope 6A |
| chrX_-_134658450 | 0.04 |
ENST00000359237.9
|
PLAC1
|
placenta enriched 1 |
| chr2_-_227724757 | 0.04 |
ENST00000641918.1
|
SCYGR6
|
small cysteine and glycine repeat containing 6 |
| chr9_-_74887720 | 0.04 |
ENST00000449912.6
|
TRPM6
|
transient receptor potential cation channel subfamily M member 6 |
| chr14_-_55191534 | 0.03 |
ENST00000395425.6
ENST00000247191.7 |
DLGAP5
|
DLG associated protein 5 |
| chr5_+_141364231 | 0.03 |
ENST00000611914.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chrX_-_139642835 | 0.03 |
ENST00000536274.5
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr1_+_33081145 | 0.03 |
ENST00000294517.11
ENST00000373443.7 |
AZIN2
|
antizyme inhibitor 2 |
| chr15_+_40953463 | 0.03 |
ENST00000617768.5
|
CHAC1
|
ChaC glutathione specific gamma-glutamylcyclotransferase 1 |
| chrX_-_139642518 | 0.02 |
ENST00000370573.8
ENST00000338585.6 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr1_-_113904789 | 0.02 |
ENST00000369569.6
ENST00000369567.5 |
AP4B1
|
adaptor related protein complex 4 subunit beta 1 |
| chr17_-_79993634 | 0.02 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family member 16 |
| chr2_-_158456702 | 0.02 |
ENST00000409889.1
ENST00000283233.10 |
CCDC148
|
coiled-coil domain containing 148 |
| chrX_-_139642889 | 0.01 |
ENST00000370576.9
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr8_+_26577843 | 0.01 |
ENST00000311151.9
|
DPYSL2
|
dihydropyrimidinase like 2 |
| chr7_-_138679045 | 0.01 |
ENST00000419765.4
|
SVOPL
|
SVOP like |
| chrX_-_100874209 | 0.00 |
ENST00000372964.5
ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr12_-_109309278 | 0.00 |
ENST00000299162.10
|
FOXN4
|
forkhead box N4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DDIT3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.5 | 4.7 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.4 | 3.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.3 | 7.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.2 | 3.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 0.9 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.1 | 2.9 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.6 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.3 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.1 | 0.6 | GO:1904640 | response to methionine(GO:1904640) |
| 0.1 | 1.1 | GO:2000334 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.2 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.1 | 0.2 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.1 | 1.2 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 1.2 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.9 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.2 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.3 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.3 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 0.5 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 0.3 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 8.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.0 | 1.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 2.1 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.5 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.6 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.0 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.3 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 2.4 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.0 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 3.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 5.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.3 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.1 | 1.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 7.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 4.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 7.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.9 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 2.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.8 | GO:0043209 | myelin sheath(GO:0043209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 1.2 | 3.7 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 1.0 | 3.0 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.3 | 5.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.3 | 4.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 2.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 3.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.9 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 1.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 1.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.1 | 0.2 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 2.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 1.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 1.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.7 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 5.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 5.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 7.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |