Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for DLX4_HOXD8
Z-value: 2.00
Transcription factors associated with DLX4_HOXD8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
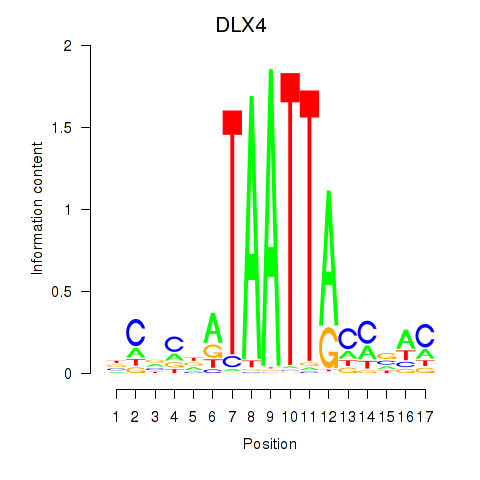
DLX4
|
ENSG00000108813.11 | distal-less homeobox 4 |
|
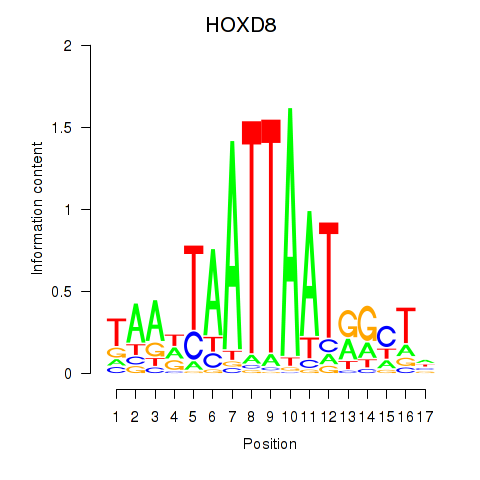
HOXD8
|
ENSG00000175879.9 | homeobox D8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD8 | hg38_v1_chr2_+_176129680_176129764 | -0.36 | 4.3e-02 | Click! |
| DLX4 | hg38_v1_chr17_+_49969178_49969214 | -0.17 | 3.4e-01 | Click! |
Activity profile of DLX4_HOXD8 motif
Sorted Z-values of DLX4_HOXD8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_157488829 | 13.41 |
ENST00000435117.1
ENST00000439355.5 |
CYTIP
|
cytohesin 1 interacting protein |
| chr14_-_106038355 | 11.52 |
ENST00000390597.3
|
IGHV2-5
|
immunoglobulin heavy variable 2-5 |
| chr7_+_142791635 | 10.61 |
ENST00000633705.1
|
TRBC1
|
T cell receptor beta constant 1 |
| chr2_+_90172802 | 10.57 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr1_-_183569186 | 9.89 |
ENST00000420553.5
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr12_+_9827472 | 9.82 |
ENST00000617793.4
ENST00000617889.5 ENST00000354855.7 ENST00000279545.7 |
KLRF1
|
killer cell lectin like receptor F1 |
| chr19_-_54313074 | 9.39 |
ENST00000486742.2
ENST00000432233.8 |
LILRA5
|
leukocyte immunoglobulin like receptor A5 |
| chr2_+_90038848 | 8.57 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr2_-_89100352 | 8.15 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr15_+_58138368 | 7.80 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chr4_-_73988179 | 7.71 |
ENST00000296028.4
|
PPBP
|
pro-platelet basic protein |
| chr2_+_90100235 | 7.70 |
ENST00000492446.1
|
IGKV1D-16
|
immunoglobulin kappa variable 1D-16 |
| chr17_-_66229380 | 7.42 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr2_-_89117844 | 7.26 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr7_+_142300924 | 7.08 |
ENST00000455382.2
|
TRBV2
|
T cell receptor beta variable 2 |
| chr14_-_24609660 | 6.80 |
ENST00000557220.6
ENST00000216338.9 ENST00000382548.4 |
GZMH
|
granzyme H |
| chr14_-_106005574 | 6.71 |
ENST00000390595.3
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr11_-_60183011 | 6.65 |
ENST00000533023.5
ENST00000420732.6 ENST00000528851.6 |
MS4A6A
|
membrane spanning 4-domains A6A |
| chr1_-_113871665 | 6.35 |
ENST00000528414.5
ENST00000460620.5 ENST00000359785.10 ENST00000420377.6 ENST00000525799.1 ENST00000538253.5 |
PTPN22
|
protein tyrosine phosphatase non-receptor type 22 |
| chr2_-_88947820 | 6.08 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chrX_+_78945332 | 6.07 |
ENST00000544091.1
ENST00000171757.3 |
P2RY10
|
P2Y receptor family member 10 |
| chr13_-_46168495 | 5.95 |
ENST00000416500.5
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr17_-_31318818 | 5.79 |
ENST00000578584.5
|
ENSG00000265118.5
|
novel protein |
| chr2_-_89085787 | 5.79 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr22_+_22711689 | 5.76 |
ENST00000390308.2
|
IGLV3-21
|
immunoglobulin lambda variable 3-21 |
| chr11_-_18236795 | 5.72 |
ENST00000278222.7
|
SAA4
|
serum amyloid A4, constitutive |
| chr2_+_87338511 | 5.62 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr1_-_160523204 | 5.43 |
ENST00000368055.1
ENST00000368057.8 ENST00000368059.7 |
SLAMF6
|
SLAM family member 6 |
| chr1_+_160739286 | 5.42 |
ENST00000359331.8
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr2_+_90234809 | 5.42 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr4_+_40196907 | 5.35 |
ENST00000622175.4
ENST00000619474.4 ENST00000615083.4 ENST00000610353.4 ENST00000614836.1 |
RHOH
|
ras homolog family member H |
| chr5_+_58491427 | 5.34 |
ENST00000396776.6
ENST00000502276.6 ENST00000511930.2 |
GAPT
|
GRB2 binding adaptor protein, transmembrane |
| chr14_-_106277039 | 5.23 |
ENST00000390610.2
|
IGHV1-24
|
immunoglobulin heavy variable 1-24 |
| chr5_+_58491451 | 5.17 |
ENST00000513924.2
ENST00000515443.2 |
GAPT
|
GRB2 binding adaptor protein, transmembrane |
| chr12_+_9827517 | 5.12 |
ENST00000537723.5
|
KLRF1
|
killer cell lectin like receptor F1 |
| chr2_+_90159840 | 5.10 |
ENST00000377032.5
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr14_-_106269133 | 5.04 |
ENST00000390609.3
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr21_-_14546297 | 5.04 |
ENST00000400566.6
ENST00000400564.5 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr14_-_106593319 | 5.03 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr3_+_149474688 | 5.01 |
ENST00000305354.5
ENST00000465758.1 |
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr4_-_39032343 | 4.89 |
ENST00000381938.4
|
TMEM156
|
transmembrane protein 156 |
| chr2_+_89913982 | 4.89 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr2_-_88857582 | 4.80 |
ENST00000390237.2
|
IGKC
|
immunoglobulin kappa constant |
| chr1_+_160739265 | 4.73 |
ENST00000368042.7
|
SLAMF7
|
SLAM family member 7 |
| chr2_+_90004792 | 4.66 |
ENST00000462693.1
|
IGKV2D-24
|
immunoglobulin kappa variable 2D-24 (non-functional) |
| chr14_+_22040576 | 4.59 |
ENST00000390448.3
|
TRAV20
|
T cell receptor alpha variable 20 |
| chr2_-_88979016 | 4.53 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr14_+_22112280 | 4.53 |
ENST00000390454.2
|
TRAV25
|
T cell receptor alpha variable 25 |
| chr12_+_9669735 | 4.48 |
ENST00000545918.5
ENST00000543300.5 ENST00000261339.10 ENST00000466035.6 |
CLEC2D
|
C-type lectin domain family 2 member D |
| chr14_+_22163226 | 4.47 |
ENST00000390458.3
|
TRAV29DV5
|
T cell receptor alpha variable 29/delta variable 5 |
| chr14_+_21868822 | 4.47 |
ENST00000390436.2
|
TRAV13-1
|
T cell receptor alpha variable 13-1 |
| chr22_+_38957522 | 4.47 |
ENST00000618553.1
ENST00000249116.7 |
APOBEC3A
|
apolipoprotein B mRNA editing enzyme catalytic subunit 3A |
| chr12_+_20810698 | 4.46 |
ENST00000540853.5
ENST00000381545.8 |
SLCO1B3
|
solute carrier organic anion transporter family member 1B3 |
| chr14_-_106470788 | 4.46 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chrX_-_30577759 | 4.41 |
ENST00000378962.4
|
TASL
|
TLR adaptor interacting with endolysosomal SLC15A4 |
| chr2_-_181680490 | 4.37 |
ENST00000684145.1
ENST00000295108.4 ENST00000684079.1 ENST00000683430.1 |
CERKL
NEUROD1
|
ceramide kinase like neuronal differentiation 1 |
| chr15_+_88635626 | 4.36 |
ENST00000379224.10
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr1_-_56966133 | 4.36 |
ENST00000535057.5
ENST00000543257.5 |
C8B
|
complement C8 beta chain |
| chr6_-_36547400 | 4.33 |
ENST00000229812.8
|
STK38
|
serine/threonine kinase 38 |
| chr22_+_22322452 | 4.32 |
ENST00000390290.3
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr10_+_7703340 | 4.30 |
ENST00000429820.5
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr9_-_114074969 | 4.28 |
ENST00000466610.6
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr1_+_160739239 | 4.24 |
ENST00000368043.8
|
SLAMF7
|
SLAM family member 7 |
| chr20_-_1657714 | 4.18 |
ENST00000216927.4
ENST00000344103.8 |
SIRPG
|
signal regulatory protein gamma |
| chr13_-_46211841 | 4.18 |
ENST00000442275.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr2_-_89213917 | 4.17 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr14_-_106185387 | 4.12 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18 |
| chr2_+_89851723 | 4.08 |
ENST00000429992.2
|
IGKV2D-40
|
immunoglobulin kappa variable 2D-40 |
| chr2_+_88885397 | 4.05 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr7_-_38249572 | 4.05 |
ENST00000436911.6
|
TRGC2
|
T cell receptor gamma constant 2 |
| chr1_+_198639162 | 4.00 |
ENST00000418674.1
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr12_+_21131187 | 3.99 |
ENST00000256958.3
|
SLCO1B1
|
solute carrier organic anion transporter family member 1B1 |
| chr14_-_106724093 | 3.94 |
ENST00000390634.3
|
IGHV2-70D
|
immunoglobulin heavy variable 2-70D |
| chr11_-_60183191 | 3.93 |
ENST00000412309.6
|
MS4A6A
|
membrane spanning 4-domains A6A |
| chr6_-_132734692 | 3.93 |
ENST00000509351.5
ENST00000417437.6 ENST00000423615.6 ENST00000427187.6 ENST00000414302.7 ENST00000367927.9 ENST00000450865.2 |
VNN3
|
vanin 3 |
| chr3_+_108822759 | 3.90 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr17_-_35880350 | 3.83 |
ENST00000605140.6
ENST00000651122.1 ENST00000603197.6 |
CCL5
|
C-C motif chemokine ligand 5 |
| chr11_+_35180279 | 3.83 |
ENST00000531873.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr3_+_46370854 | 3.83 |
ENST00000292303.4
|
CCR5
|
C-C motif chemokine receptor 5 |
| chr5_+_55102635 | 3.82 |
ENST00000274306.7
|
GZMA
|
granzyme A |
| chr4_+_73740541 | 3.81 |
ENST00000401931.1
ENST00000307407.8 |
CXCL8
|
C-X-C motif chemokine ligand 8 |
| chr1_+_198638457 | 3.80 |
ENST00000367379.6
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr1_-_92486916 | 3.77 |
ENST00000294702.6
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr12_-_9607903 | 3.76 |
ENST00000229402.4
|
KLRB1
|
killer cell lectin like receptor B1 |
| chr2_+_90114838 | 3.73 |
ENST00000417279.3
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 |
| chr2_+_90220727 | 3.70 |
ENST00000471857.2
|
IGKV1D-8
|
immunoglobulin kappa variable 1D-8 |
| chr1_+_158831323 | 3.70 |
ENST00000368141.5
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr15_-_79971164 | 3.67 |
ENST00000335661.6
ENST00000267953.4 ENST00000677151.1 |
BCL2A1
|
BCL2 related protein A1 |
| chr1_+_196774813 | 3.64 |
ENST00000471440.6
ENST00000391985.7 ENST00000617219.1 ENST00000367425.9 |
CFHR3
|
complement factor H related 3 |
| chr4_-_38804783 | 3.51 |
ENST00000308979.7
ENST00000505940.1 ENST00000515861.5 |
TLR1
|
toll like receptor 1 |
| chr14_+_20955484 | 3.49 |
ENST00000304625.3
|
RNASE2
|
ribonuclease A family member 2 |
| chr19_+_14583076 | 3.48 |
ENST00000547437.5
ENST00000417570.6 |
CLEC17A
|
C-type lectin domain containing 17A |
| chr14_-_106130061 | 3.48 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr10_+_94683771 | 3.45 |
ENST00000339022.6
|
CYP2C18
|
cytochrome P450 family 2 subfamily C member 18 |
| chr1_-_160579439 | 3.44 |
ENST00000368054.8
ENST00000368048.7 ENST00000311224.8 ENST00000368051.3 ENST00000534968.5 |
CD84
|
CD84 molecule |
| chr14_-_106511856 | 3.43 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr2_-_218166951 | 3.42 |
ENST00000295683.3
|
CXCR1
|
C-X-C motif chemokine receptor 1 |
| chr7_-_116030750 | 3.41 |
ENST00000265440.12
ENST00000320239.11 |
TFEC
|
transcription factor EC |
| chr12_-_10409757 | 3.40 |
ENST00000309384.2
|
KLRC4
|
killer cell lectin like receptor C4 |
| chr11_-_118225086 | 3.39 |
ENST00000640745.1
|
JAML
|
junction adhesion molecule like |
| chr2_-_89143133 | 3.38 |
ENST00000492167.1
|
IGKV3-20
|
immunoglobulin kappa variable 3-20 |
| chr1_+_158931539 | 3.37 |
ENST00000368140.6
ENST00000368138.7 ENST00000392254.6 ENST00000392252.7 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family member 1 |
| chr7_+_142450941 | 3.36 |
ENST00000390368.2
|
TRBV6-5
|
T cell receptor beta variable 6-5 |
| chr14_-_106335613 | 3.36 |
ENST00000603660.1
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr17_+_36103819 | 3.36 |
ENST00000615863.2
ENST00000621626.1 |
CCL4
|
C-C motif chemokine ligand 4 |
| chr4_-_25863537 | 3.36 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr4_-_70666492 | 3.35 |
ENST00000254801.9
ENST00000391614.7 |
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr21_-_14658812 | 3.35 |
ENST00000647101.1
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr17_+_36211055 | 3.35 |
ENST00000617405.5
ENST00000617416.4 ENST00000613173.4 ENST00000620732.4 ENST00000620098.4 ENST00000620576.4 ENST00000620055.4 ENST00000610565.4 ENST00000620250.1 |
CCL4L2
|
C-C motif chemokine ligand 4 like 2 |
| chr7_+_142492121 | 3.35 |
ENST00000390374.3
|
TRBV7-6
|
T cell receptor beta variable 7-6 |
| chr1_+_206897435 | 3.31 |
ENST00000391929.7
ENST00000294984.7 ENST00000611909.4 ENST00000367093.3 |
IL24
|
interleukin 24 |
| chr4_+_40197023 | 3.30 |
ENST00000381799.10
|
RHOH
|
ras homolog family member H |
| chr20_+_24949256 | 3.28 |
ENST00000480798.2
|
CST7
|
cystatin F |
| chr4_-_154590735 | 3.26 |
ENST00000403106.8
ENST00000622532.1 ENST00000651975.1 |
FGA
|
fibrinogen alpha chain |
| chr10_+_7703300 | 3.18 |
ENST00000358415.9
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr12_-_14961610 | 3.18 |
ENST00000542276.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr12_-_10130143 | 3.15 |
ENST00000298523.9
ENST00000396484.6 ENST00000310002.4 ENST00000304084.13 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr4_-_68670648 | 3.08 |
ENST00000338206.6
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15 |
| chr21_-_14546351 | 3.08 |
ENST00000619120.4
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr7_+_150685693 | 3.07 |
ENST00000223293.10
ENST00000474605.1 |
GIMAP2
|
GTPase, IMAP family member 2 |
| chr6_-_132763424 | 3.06 |
ENST00000532012.1
ENST00000525270.5 ENST00000530536.5 ENST00000524919.5 |
VNN2
|
vanin 2 |
| chr19_+_51761167 | 3.05 |
ENST00000340023.7
ENST00000599326.1 ENST00000598953.1 |
FPR2
|
formyl peptide receptor 2 |
| chr4_-_48080172 | 3.04 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr12_-_10130241 | 3.03 |
ENST00000353231.9
ENST00000525605.1 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr12_-_14950606 | 3.03 |
ENST00000536592.5
|
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr11_-_118225002 | 3.03 |
ENST00000356289.10
ENST00000526620.5 |
JAML
|
junction adhesion molecule like |
| chr22_+_22811737 | 3.00 |
ENST00000390315.3
|
IGLV3-10
|
immunoglobulin lambda variable 3-10 |
| chr2_-_88861920 | 2.99 |
ENST00000390242.2
|
IGKJ1
|
immunoglobulin kappa joining 1 |
| chr15_+_94355956 | 2.99 |
ENST00000557742.1
|
MCTP2
|
multiple C2 and transmembrane domain containing 2 |
| chr2_+_102418642 | 2.99 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr2_+_191678967 | 2.98 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr6_-_127918604 | 2.98 |
ENST00000537166.5
|
THEMIS
|
thymocyte selection associated |
| chr3_-_39280021 | 2.96 |
ENST00000399220.3
|
CX3CR1
|
C-X3-C motif chemokine receptor 1 |
| chr12_-_9999176 | 2.95 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr17_-_66220630 | 2.93 |
ENST00000585162.1
|
APOH
|
apolipoprotein H |
| chr2_+_203936755 | 2.93 |
ENST00000316386.11
ENST00000435193.1 |
ICOS
|
inducible T cell costimulator |
| chr14_-_106771020 | 2.92 |
ENST00000617374.2
|
IGHV2-70
|
immunoglobulin heavy variable 2-70 |
| chr5_+_76875177 | 2.92 |
ENST00000613039.1
|
S100Z
|
S100 calcium binding protein Z |
| chr22_+_44180915 | 2.91 |
ENST00000444313.8
ENST00000416291.5 |
PARVG
|
parvin gamma |
| chr2_-_157444044 | 2.90 |
ENST00000264192.8
|
CYTIP
|
cytohesin 1 interacting protein |
| chr1_+_56854764 | 2.88 |
ENST00000361249.4
|
C8A
|
complement C8 alpha chain |
| chr6_-_132714045 | 2.87 |
ENST00000367928.5
|
VNN1
|
vanin 1 |
| chr1_-_56966006 | 2.82 |
ENST00000371237.9
|
C8B
|
complement C8 beta chain |
| chr1_+_196977550 | 2.82 |
ENST00000256785.5
|
CFHR5
|
complement factor H related 5 |
| chr22_+_22395005 | 2.81 |
ENST00000390298.2
|
IGLV7-43
|
immunoglobulin lambda variable 7-43 |
| chr7_-_116030735 | 2.80 |
ENST00000393485.5
|
TFEC
|
transcription factor EC |
| chr2_-_136118142 | 2.78 |
ENST00000241393.4
|
CXCR4
|
C-X-C motif chemokine receptor 4 |
| chr2_+_90209873 | 2.77 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chrX_+_37780049 | 2.76 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain |
| chr19_+_49513353 | 2.74 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr2_+_89936859 | 2.74 |
ENST00000474213.1
|
IGKV2D-30
|
immunoglobulin kappa variable 2D-30 |
| chr19_-_14674829 | 2.74 |
ENST00000443157.6
ENST00000253673.6 |
ADGRE3
|
adhesion G protein-coupled receptor E3 |
| chr14_-_106360320 | 2.72 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
| chr2_-_89040745 | 2.71 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr2_+_218125276 | 2.67 |
ENST00000453237.5
|
CXCR2
|
C-X-C motif chemokine receptor 2 |
| chr2_-_159798234 | 2.67 |
ENST00000429078.6
ENST00000553424.5 |
CD302
|
CD302 molecule |
| chr7_-_38349972 | 2.66 |
ENST00000390344.2
|
TRGV5
|
T cell receptor gamma variable 5 |
| chr2_-_159798043 | 2.65 |
ENST00000664982.1
ENST00000259053.6 |
ENSG00000287091.1
CD302
|
novel transcript, sense intronic to CD302and LY75-CD302 CD302 molecule |
| chr22_+_22380766 | 2.64 |
ENST00000390297.3
|
IGLV1-44
|
immunoglobulin lambda variable 1-44 |
| chr16_-_28623560 | 2.63 |
ENST00000350842.8
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chr5_-_135954962 | 2.62 |
ENST00000522943.5
ENST00000514447.2 ENST00000274507.6 |
LECT2
|
leukocyte cell derived chemotaxin 2 |
| chr4_+_68815991 | 2.62 |
ENST00000265403.12
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase family 2 member B10 |
| chr16_-_71577082 | 2.61 |
ENST00000355962.5
|
TAT
|
tyrosine aminotransferase |
| chr4_+_99816797 | 2.61 |
ENST00000512369.2
ENST00000296414.11 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides 1 |
| chr16_-_21652598 | 2.60 |
ENST00000569602.1
ENST00000268389.6 |
IGSF6
|
immunoglobulin superfamily member 6 |
| chr17_+_69502397 | 2.59 |
ENST00000613873.4
ENST00000589647.5 |
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr8_-_85341659 | 2.58 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr17_-_445939 | 2.56 |
ENST00000329099.4
|
RFLNB
|
refilin B |
| chr4_+_154563003 | 2.55 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr8_-_6978634 | 2.55 |
ENST00000382679.2
|
DEFA1
|
defensin alpha 1 |
| chr6_+_131573219 | 2.54 |
ENST00000356962.2
ENST00000368087.8 ENST00000673427.1 ENST00000640973.1 |
ARG1
|
arginase 1 |
| chr2_-_89027700 | 2.53 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11 |
| chr14_+_21965451 | 2.52 |
ENST00000390442.3
|
TRAV12-3
|
T cell receptor alpha variable 12-3 |
| chr22_+_22409755 | 2.51 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr7_+_80626148 | 2.49 |
ENST00000428497.5
|
CD36
|
CD36 molecule |
| chr3_+_186974957 | 2.49 |
ENST00000438590.5
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr7_-_36724380 | 2.48 |
ENST00000617267.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr14_+_21924033 | 2.48 |
ENST00000390440.2
|
TRAV14DV4
|
T cell receptor alpha variable 14/delta variable 4 |
| chr6_-_132757883 | 2.48 |
ENST00000525289.5
ENST00000326499.11 |
VNN2
|
vanin 2 |
| chr22_+_22162155 | 2.47 |
ENST00000390284.2
|
IGLV4-60
|
immunoglobulin lambda variable 4-60 |
| chr3_+_108822778 | 2.45 |
ENST00000295756.11
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr2_+_218125671 | 2.45 |
ENST00000415392.5
|
CXCR2
|
C-X-C motif chemokine receptor 2 |
| chr19_-_58353482 | 2.43 |
ENST00000263100.8
|
A1BG
|
alpha-1-B glycoprotein |
| chr3_+_32391841 | 2.42 |
ENST00000334983.10
|
CMTM7
|
CKLF like MARVEL transmembrane domain containing 7 |
| chr11_+_60280658 | 2.41 |
ENST00000337908.5
|
MS4A4A
|
membrane spanning 4-domains A4A |
| chr3_-_194351290 | 2.41 |
ENST00000429275.1
ENST00000323830.4 |
CPN2
|
carboxypeptidase N subunit 2 |
| chr6_-_127918540 | 2.40 |
ENST00000368250.5
|
THEMIS
|
thymocyte selection associated |
| chr6_-_32941018 | 2.39 |
ENST00000418107.3
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr7_+_80624961 | 2.39 |
ENST00000436384.5
|
CD36
|
CD36 molecule |
| chr14_-_91253925 | 2.39 |
ENST00000531499.2
|
GPR68
|
G protein-coupled receptor 68 |
| chr1_+_196943738 | 2.39 |
ENST00000367415.8
ENST00000367421.5 ENST00000649283.1 ENST00000476712.6 ENST00000496448.6 ENST00000473386.1 ENST00000649960.1 |
CFHR2
|
complement factor H related 2 |
| chr1_-_183590876 | 2.37 |
ENST00000367536.5
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr20_+_3786772 | 2.35 |
ENST00000344256.10
ENST00000379598.9 |
CDC25B
|
cell division cycle 25B |
| chr4_+_73409340 | 2.35 |
ENST00000511370.1
|
ALB
|
albumin |
| chr4_-_48114523 | 2.35 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr7_-_77199808 | 2.35 |
ENST00000248598.6
|
FGL2
|
fibrinogen like 2 |
| chr1_+_198638968 | 2.33 |
ENST00000348564.11
ENST00000530727.5 ENST00000442510.8 ENST00000645247.1 ENST00000367367.8 ENST00000367364.5 ENST00000413409.6 |
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr2_+_201183120 | 2.28 |
ENST00000272879.9
ENST00000286186.11 ENST00000374650.7 ENST00000346817.9 ENST00000313728.11 ENST00000448480.1 |
CASP10
|
caspase 10 |
| chr7_+_76461676 | 2.28 |
ENST00000425780.5
ENST00000456590.5 ENST00000451769.5 ENST00000324432.9 ENST00000457529.5 ENST00000446600.5 ENST00000430490.7 ENST00000413936.6 ENST00000423646.5 ENST00000438930.5 |
DTX2
|
deltex E3 ubiquitin ligase 2 |
| chr7_-_36724457 | 2.28 |
ENST00000617537.5
ENST00000435386.1 |
AOAH
|
acyloxyacyl hydrolase |
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX4_HOXD8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 12.3 | GO:2000473 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 1.7 | 6.7 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 1.7 | 9.9 | GO:0038112 | interleukin-8-mediated signaling pathway(GO:0038112) |
| 1.7 | 5.0 | GO:0034130 | toll-like receptor 1 signaling pathway(GO:0034130) |
| 1.6 | 10.9 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 1.5 | 7.7 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 1.5 | 4.4 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 1.4 | 4.3 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 1.3 | 6.5 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 1.3 | 3.9 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 1.3 | 22.7 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 1.3 | 3.8 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 1.2 | 204.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 1.2 | 7.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.2 | 4.8 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 1.2 | 2.4 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 1.2 | 7.0 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 1.1 | 3.4 | GO:0032701 | negative regulation of interleukin-18 production(GO:0032701) |
| 1.1 | 4.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.9 | 2.8 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.9 | 2.7 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.9 | 6.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.9 | 6.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.9 | 2.6 | GO:0070409 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.9 | 3.4 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.9 | 2.6 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.8 | 2.5 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.8 | 6.7 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.8 | 2.5 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.8 | 4.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.8 | 2.3 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.8 | 10.7 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.7 | 2.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.7 | 2.9 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.7 | 12.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.7 | 10.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.7 | 3.6 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.7 | 2.1 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.7 | 2.7 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.7 | 6.6 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.7 | 2.0 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.6 | 3.2 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.6 | 3.8 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.6 | 2.5 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.6 | 3.1 | GO:0038124 | toll-like receptor TLR6:TLR2 signaling pathway(GO:0038124) response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.6 | 3.7 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.6 | 1.8 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.6 | 1.8 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.6 | 4.0 | GO:0072011 | glomerular endothelium development(GO:0072011) |
| 0.6 | 2.3 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.5 | 1.6 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.5 | 4.3 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.5 | 1.6 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.5 | 1.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.5 | 7.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.5 | 1.5 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.5 | 2.0 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.5 | 1.5 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.5 | 2.0 | GO:0045401 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) |
| 0.5 | 1.9 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.5 | 0.9 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 0.5 | 12.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.5 | 6.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.5 | 17.6 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.5 | 1.4 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.4 | 1.8 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.4 | 1.7 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.4 | 1.3 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.4 | 1.3 | GO:2000469 | regulation of thioredoxin peroxidase activity(GO:1903123) negative regulation of thioredoxin peroxidase activity(GO:1903124) negative regulation of thioredoxin peroxidase activity by peptidyl-threonine phosphorylation(GO:1903125) Wnt signalosome assembly(GO:1904887) negative regulation of peroxidase activity(GO:2000469) |
| 0.4 | 12.0 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.4 | 10.7 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.4 | 0.4 | GO:0046645 | positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.4 | 0.8 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.4 | 8.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.4 | 1.2 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.4 | 3.9 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.4 | 1.5 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.4 | 5.0 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.4 | 1.5 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.4 | 13.1 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.4 | 1.1 | GO:0032831 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.4 | 9.4 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.4 | 2.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.4 | 1.1 | GO:0016107 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.4 | 8.8 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.3 | 2.4 | GO:0090096 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.3 | 4.9 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.3 | 1.0 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.3 | 2.7 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.3 | 1.0 | GO:0060003 | copper ion export(GO:0060003) |
| 0.3 | 1.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.3 | 1.0 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.3 | 1.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.3 | 5.9 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.3 | 1.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.3 | 1.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.3 | 1.3 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.3 | 6.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 0.6 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.3 | 2.9 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.3 | 0.6 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.3 | 0.9 | GO:1902688 | regulation of NAD metabolic process(GO:1902688) |
| 0.3 | 2.4 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.3 | 0.6 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.3 | 60.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.3 | 0.9 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.3 | 0.9 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.3 | 1.4 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.3 | 1.6 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.3 | 3.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.3 | 1.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 0.8 | GO:1903414 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) spleen trabecula formation(GO:0060345) divalent metal ion export(GO:0070839) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.3 | 2.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.3 | 1.3 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.3 | 5.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 3.8 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.2 | 1.0 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 1.7 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.2 | 0.7 | GO:1904640 | response to methionine(GO:1904640) |
| 0.2 | 0.7 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 1.9 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 3.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 0.7 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.2 | 0.5 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 | 1.4 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.2 | 2.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 1.7 | GO:0050955 | thermoception(GO:0050955) |
| 0.2 | 1.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.2 | 1.7 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 | 2.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 0.6 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.2 | 1.4 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.2 | 1.4 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.2 | 1.2 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.2 | 1.0 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) |
| 0.2 | 2.8 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 | 1.4 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.2 | 0.6 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.2 | 1.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.2 | 0.8 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.2 | 1.1 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.2 | 15.2 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.2 | 0.8 | GO:0090260 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.2 | 0.6 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.2 | 0.6 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.2 | 2.6 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.2 | 0.7 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.2 | 1.8 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 0.7 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.2 | 0.5 | GO:1902960 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) regulation of neurofibrillary tangle assembly(GO:1902996) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.2 | 1.2 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.2 | 0.5 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.2 | 4.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 0.3 | GO:0051709 | regulation of killing of cells of other organism(GO:0051709) positive regulation of killing of cells of other organism(GO:0051712) |
| 0.2 | 0.5 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 0.2 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.2 | 0.5 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.2 | 0.9 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.2 | 4.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 0.8 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 0.6 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.2 | 0.8 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.2 | 0.8 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.2 | 1.4 | GO:0070942 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.2 | 1.2 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.2 | 0.9 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.2 | 0.3 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 | 12.7 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.4 | GO:0045416 | positive regulation of interleukin-8 biosynthetic process(GO:0045416) |
| 0.1 | 1.0 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.7 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 0.4 | GO:0090340 | positive regulation of high-density lipoprotein particle assembly(GO:0090108) positive regulation of secretion of lysosomal enzymes(GO:0090340) |
| 0.1 | 1.4 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 1.0 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.7 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 0.3 | GO:0033029 | neutrophil apoptotic process(GO:0001781) regulation of neutrophil apoptotic process(GO:0033029) |
| 0.1 | 0.5 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.7 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.1 | 7.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.4 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.1 | 1.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.8 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 5.4 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.1 | 1.2 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) |
| 0.1 | 0.4 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 8.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 2.4 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 2.4 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 1.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.6 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.1 | 0.6 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.7 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 1.3 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 1.0 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 1.3 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.6 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 2.5 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.1 | 0.9 | GO:0006203 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.1 | 0.4 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) RNA repair(GO:0042245) |
| 0.1 | 0.8 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.9 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.5 | GO:0044010 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.1 | 0.8 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.1 | 5.0 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.1 | 0.9 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 | 0.7 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 2.1 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.1 | 0.7 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.1 | 1.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.4 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 1.4 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.4 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.6 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 1.0 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.3 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.1 | 0.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 2.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 6.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 1.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.5 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.6 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.4 | GO:0044035 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.6 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 2.5 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 1.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.3 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.1 | 1.7 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.1 | 0.5 | GO:0070378 | positive regulation of ERK5 cascade(GO:0070378) |
| 0.1 | 1.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.5 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) |
| 0.1 | 0.1 | GO:1903006 | regulation of protein K63-linked deubiquitination(GO:1903004) positive regulation of protein K63-linked deubiquitination(GO:1903006) |
| 0.1 | 0.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.8 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.1 | 1.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.5 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 3.5 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.5 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.3 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 2.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.6 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.1 | 0.3 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 4.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 2.0 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 0.4 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.3 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.2 | GO:0045799 | regulation of heterochromatin assembly(GO:0031445) positive regulation of heterochromatin assembly(GO:0031453) positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.1 | 0.4 | GO:0043375 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 | 1.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.3 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 1.3 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.1 | 0.4 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 0.4 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.1 | 0.4 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.6 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.3 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.1 | 0.3 | GO:0072560 | type B pancreatic cell maturation(GO:0072560) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.1 | 10.4 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 0.8 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.8 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.5 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.3 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.1 | 0.5 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.6 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 1.6 | GO:1904037 | positive regulation of epithelial cell apoptotic process(GO:1904037) |
| 0.1 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.1 | 0.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 1.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 10.3 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 1.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 1.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 0.1 | GO:0046060 | dATP metabolic process(GO:0046060) |
| 0.1 | 0.9 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.1 | 1.2 | GO:0042346 | positive regulation of NF-kappaB import into nucleus(GO:0042346) |
| 0.1 | 0.3 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.1 | 0.8 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 0.8 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 0.8 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 2.2 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.1 | 0.6 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 2.3 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 1.9 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.5 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.2 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 0.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.4 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.8 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.2 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.1 | 0.3 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 1.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.7 | GO:1903944 | negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.4 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.4 | GO:0001519 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.1 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.2 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 0.2 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 0.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.2 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.1 | 0.5 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 1.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.9 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 0.6 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 3.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 2.5 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:0098503 | nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) DNA 3' dephosphorylation(GO:0098503) DNA 3' dephosphorylation involved in DNA repair(GO:0098504) polynucleotide 3' dephosphorylation(GO:0098506) |
| 0.0 | 0.2 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.4 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.4 | GO:1903961 | positive regulation of anion channel activity(GO:1901529) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 1.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.0 | 0.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.8 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.2 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.8 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.0 | 0.5 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.8 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 16.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.4 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.3 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.2 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.8 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 1.8 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.2 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.7 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.2 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.2 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.5 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 1.0 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.9 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.5 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.3 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.6 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 2.4 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.4 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.2 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 5.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.6 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.8 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 21.9 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.7 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 1.3 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) telomere maintenance via base-excision repair(GO:0097698) DNA dephosphorylation(GO:0098502) |
| 0.0 | 4.5 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 1.3 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 0.0 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 1.2 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.0 | 0.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.2 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:1904732 | regulation of electron carrier activity(GO:1904732) |
| 0.0 | 0.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.2 | GO:0072526 | purine ribonucleoside salvage(GO:0006166) L-methionine biosynthetic process from methylthioadenosine(GO:0019509) pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.8 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 4.3 | GO:0050870 | positive regulation of T cell activation(GO:0050870) |
| 0.0 | 0.8 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.4 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 1.6 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.7 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.5 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 1.8 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:0071442 | regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.9 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.4 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.2 | GO:0010816 | neuropeptide catabolic process(GO:0010813) substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.4 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.6 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.1 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 | 2.4 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.7 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 1.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 1.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.6 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.0 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) organ senescence(GO:0010260) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 3.0 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.5 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.3 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.9 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.5 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 2.1 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 2.0 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.4 | GO:0050715 | positive regulation of cytokine secretion(GO:0050715) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:1990668 | vesicle fusion with endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membrane(GO:1990668) |
| 0.0 | 1.2 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.0 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:1990592 | protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.5 | GO:1902603 | carnitine transmembrane transport(GO:1902603) |
| 0.0 | 2.9 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.2 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.2 | GO:2000618 | regulation of histone H4-K16 acetylation(GO:2000618) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.4 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:1901731 | positive regulation of platelet aggregation(GO:1901731) |
| 0.0 | 0.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.5 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 1.2 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.3 | GO:0033003 | regulation of mast cell activation(GO:0033003) |
| 0.0 | 0.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 11.8 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 0.0 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.3 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.2 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.4 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.3 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.7 | GO:0071754 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 1.3 | 14.3 | GO:0032010 | phagolysosome(GO:0032010) |
| 1.2 | 5.0 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 1.1 | 10.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.1 | 58.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.9 | 12.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.9 | 10.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.6 | 3.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.5 | 2.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.5 | 24.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.5 | 2.9 | GO:0097169 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 0.4 | 80.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.4 | 3.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.4 | 8.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 1.3 | GO:0032473 | cytoplasmic side of mitochondrial outer membrane(GO:0032473) caveola neck(GO:0099400) |
| 0.3 | 3.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 11.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 13.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 5.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 2.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.3 | 8.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.3 | 1.8 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.2 | 0.7 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.2 | 1.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.2 | 3.0 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 2.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 14.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 1.6 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.2 | 0.8 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.2 | 3.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 4.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.2 | 0.2 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.2 | 0.3 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.2 | 6.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 1.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.2 | 0.8 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.2 | 0.3 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 1.8 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.8 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 2.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.4 | GO:0090651 | apical cytoplasm(GO:0090651) |
| 0.1 | 1.9 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.8 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 14.7 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.1 | 10.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 18.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 0.8 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 8.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 0.7 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 40.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.8 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 8.3 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 0.4 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.1 | 0.5 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.1 | 0.7 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 5.0 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 0.4 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 0.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.9 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.6 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 1.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 7.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.3 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.4 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.1 | 1.9 | GO:0034358 | plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.1 | 0.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 4.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 0.9 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 1.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.9 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.7 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 1.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.5 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 13.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 3.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 1.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 4.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 1.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.4 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 1.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 3.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.4 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.3 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 2.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.7 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 0.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 9.3 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.3 | GO:0038201 | TORC1 complex(GO:0031931) TOR complex(GO:0038201) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 12.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 1.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 2.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.6 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 1.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.7 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.5 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.0 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 1.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 2.3 | 13.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 2.1 | 6.3 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 1.7 | 9.9 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 1.6 | 8.0 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 1.5 | 7.7 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 1.5 | 4.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.2 | 5.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 1.2 | 4.8 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 1.2 | 17.8 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 1.2 | 6.9 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 1.1 | 7.6 | GO:0019862 | IgA binding(GO:0019862) |
| 1.0 | 14.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.9 | 4.6 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.9 | 2.8 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.9 | 248.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.9 | 4.5 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.9 | 2.6 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.9 | 2.6 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.8 | 9.6 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.8 | 12.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.7 | 2.2 | GO:0052853 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.7 | 3.7 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.7 | 7.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.7 | 2.7 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.7 | 2.0 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.6 | 2.6 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.6 | 3.7 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.6 | 3.5 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.6 | 4.0 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.6 | 3.9 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.6 | 2.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.5 | 9.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.5 | 14.9 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.5 | 5.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.5 | 3.5 | GO:0042806 | fucose binding(GO:0042806) |
| 0.5 | 12.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.5 | 1.9 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.5 | 3.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.4 | 10.7 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.4 | 8.4 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.4 | 1.7 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.4 | 1.3 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) peroxidase inhibitor activity(GO:0036479) |
| 0.4 | 13.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.4 | 8.1 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.4 | 1.9 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.4 | 1.1 | GO:0052815 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.4 | 1.4 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.4 | 5.4 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.4 | 1.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.3 | 1.0 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.3 | 1.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.3 | 1.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.3 | 1.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.3 | 2.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.3 | 0.8 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.3 | 2.8 | GO:0019863 | IgE binding(GO:0019863) |
| 0.3 | 13.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 0.8 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.3 | 0.8 | GO:0032428 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.3 | 1.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 2.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.2 | 2.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 0.7 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.2 | 1.7 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 0.7 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.2 | 11.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.5 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 1.4 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.2 | 1.4 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.2 | 10.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 0.9 | GO:0008832 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) guanyl deoxyribonucleotide binding(GO:0032560) dGTP binding(GO:0032567) |
| 0.2 | 1.4 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.2 | 3.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 0.6 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.2 | 1.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 0.9 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 2.1 | GO:0001851 | complement component C3b binding(GO:0001851) ICAM-3 receptor activity(GO:0030369) |
| 0.2 | 1.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.2 | 0.6 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.2 | 12.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.2 | 1.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 1.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 1.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.2 | 5.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 1.7 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.2 | 2.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 1.3 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.2 | 0.7 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.2 | 1.4 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.2 | 0.5 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 2.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 1.4 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 0.9 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.2 | 0.8 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 2.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.7 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 1.2 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 0.7 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 1.5 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.4 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.1 | 1.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 2.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.8 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 1.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 2.8 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.9 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 2.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.8 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.5 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 1.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.9 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.3 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.1 | 1.0 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.3 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 1.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.6 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.1 | 0.8 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.4 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 1.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 1.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.5 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.4 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 2.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.4 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 1.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 4.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 2.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.7 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.8 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 1.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 2.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.3 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.1 | 0.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.5 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.1 | 0.5 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.1 | 0.4 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.8 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 2.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.4 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 6.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 1.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.2 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.1 | 5.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 2.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 3.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 2.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.3 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.5 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 0.3 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.6 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.8 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 2.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 3.1 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.4 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.7 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.5 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 1.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.2 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 0.6 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 0.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 2.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.3 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.4 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 0.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.5 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 0.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.5 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 1.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 2.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.7 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.3 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.4 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.8 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 20.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.5 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 8.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.0 | 0.1 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.0 | 0.4 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 1.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.4 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 11.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 10.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.5 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.0 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 3.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 6.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 4.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004577 | N-acetylglucosaminyldiphosphodolichol N-acetylglucosaminyltransferase activity(GO:0004577) |
| 0.0 | 0.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 6.8 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.7 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 1.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 1.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.4 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.6 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 1.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 2.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.3 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.3 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 1.0 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.0 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 2.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0016416 | O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 1.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.0 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:1900750 | oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.3 | 11.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 9.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 24.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.2 | 15.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.2 | 14.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 4.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 0.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.2 | 3.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 14.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 8.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 5.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 18.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 3.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 3.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 5.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 3.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 2.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 2.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 5.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 6.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 5.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 21.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.9 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 4.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.3 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 2.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 7.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.6 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.8 | 13.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.7 | 3.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.5 | 9.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.5 | 8.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 13.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.5 | 34.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 10.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 12.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.4 | 7.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 4.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 8.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.4 | 17.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 5.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 6.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 5.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.2 | 3.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 11.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 4.7 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.2 | 4.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 2.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 4.0 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.2 | 7.0 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.2 | 17.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 5.8 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 4.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 6.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 3.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 3.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 3.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 9.9 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 15.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.8 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 1.9 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |
| 0.1 | 2.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 2.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 21.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.9 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 6.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.8 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.1 | 2.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 3.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 3.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.6 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.7 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 0.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 1.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 6.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 2.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 2.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 10.2 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 9.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 4.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 1.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.7 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.0 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |