Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
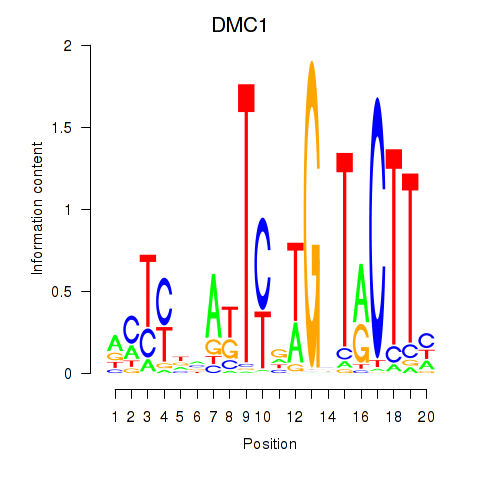
Results for DMC1
Z-value: 0.78
Transcription factors associated with DMC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DMC1
|
ENSG00000100206.10 | DNA meiotic recombinase 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DMC1 | hg38_v1_chr22_-_38570118_38570134 | 0.49 | 4.0e-03 | Click! |
Activity profile of DMC1 motif
Sorted Z-values of DMC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_95391361 | 3.43 |
ENST00000283357.10
|
FAM81B
|
family with sequence similarity 81 member B |
| chr10_+_68106109 | 3.42 |
ENST00000540630.5
ENST00000354393.6 |
MYPN
|
myopalladin |
| chr10_-_102837406 | 2.87 |
ENST00000369887.4
ENST00000638272.1 ENST00000639393.1 ENST00000638971.1 ENST00000638190.1 |
CYP17A1
|
cytochrome P450 family 17 subfamily A member 1 |
| chrX_-_50200358 | 2.68 |
ENST00000437370.2
ENST00000376064.7 ENST00000448865.5 |
AKAP4
|
A-kinase anchoring protein 4 |
| chrX_-_50200988 | 2.32 |
ENST00000358526.7
|
AKAP4
|
A-kinase anchoring protein 4 |
| chr17_+_32991844 | 2.27 |
ENST00000269053.8
|
SPACA3
|
sperm acrosome associated 3 |
| chr20_+_2816343 | 2.25 |
ENST00000380585.2
|
TMEM239
|
transmembrane protein 239 |
| chrX_+_111876177 | 2.20 |
ENST00000635763.2
|
TRPC5OS
|
TRPC5 opposite strand |
| chr1_+_166989254 | 2.07 |
ENST00000367872.9
ENST00000447624.1 |
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr2_+_170783781 | 2.01 |
ENST00000409885.1
|
ERICH2
|
glutamate rich 2 |
| chr6_+_46693835 | 1.96 |
ENST00000450697.1
|
TDRD6
|
tudor domain containing 6 |
| chr9_-_34729482 | 1.95 |
ENST00000378788.4
|
FAM205A
|
family with sequence similarity 205 member A |
| chr1_+_166989089 | 1.89 |
ENST00000367870.6
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr17_+_32991867 | 1.81 |
ENST00000394638.1
|
SPACA3
|
sperm acrosome associated 3 |
| chr2_+_119429889 | 1.67 |
ENST00000409826.1
ENST00000417645.1 |
TMEM37
|
transmembrane protein 37 |
| chr5_+_80319997 | 1.58 |
ENST00000296739.6
|
SPZ1
|
spermatogenic leucine zipper 1 |
| chrX_-_64230600 | 1.52 |
ENST00000362002.3
|
ASB12
|
ankyrin repeat and SOCS box containing 12 |
| chr7_-_5970632 | 1.52 |
ENST00000455618.2
ENST00000405415.5 ENST00000404406.5 ENST00000542644.1 |
RSPH10B
|
radial spoke head 10 homolog B |
| chr10_+_17809337 | 1.47 |
ENST00000569591.3
|
MRC1
|
mannose receptor C-type 1 |
| chrX_-_135171398 | 1.43 |
ENST00000276241.11
ENST00000344129.2 |
CT55
|
cancer/testis antigen 55 |
| chr3_+_148739798 | 1.31 |
ENST00000402260.2
|
AGTR1
|
angiotensin II receptor type 1 |
| chr20_+_2816302 | 1.28 |
ENST00000361033.1
|
TMEM239
|
transmembrane protein 239 |
| chr3_-_46812558 | 1.27 |
ENST00000641183.1
ENST00000460241.2 |
ENSG00000284672.1
ENSG00000206549.13
|
novel transcript novel protein identical to PRSS50 |
| chr7_+_90154442 | 1.26 |
ENST00000297205.7
|
STEAP1
|
STEAP family member 1 |
| chr7_+_6754109 | 1.26 |
ENST00000403107.5
ENST00000404077.5 ENST00000435395.5 ENST00000418406.1 |
RSPH10B2
|
radial spoke head 10 homolog B2 |
| chr2_+_186590022 | 1.22 |
ENST00000261023.8
ENST00000374907.7 |
ITGAV
|
integrin subunit alpha V |
| chr11_+_71527267 | 1.08 |
ENST00000398536.6
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr1_+_9539431 | 1.07 |
ENST00000302692.7
|
SLC25A33
|
solute carrier family 25 member 33 |
| chr12_-_9116223 | 1.06 |
ENST00000404455.2
|
A2M
|
alpha-2-macroglobulin |
| chr10_-_27983103 | 1.01 |
ENST00000672841.1
|
ODAD2
|
outer dynein arm docking complex subunit 2 |
| chr6_+_87344812 | 1.00 |
ENST00000388923.5
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr2_-_48755718 | 0.95 |
ENST00000294954.12
ENST00000403273.5 ENST00000401907.5 |
LHCGR
|
luteinizing hormone/choriogonadotropin receptor |
| chr11_-_74009077 | 0.93 |
ENST00000314032.9
ENST00000426995.2 |
UCP3
|
uncoupling protein 3 |
| chr12_-_120368069 | 0.93 |
ENST00000546985.1
|
MSI1
|
musashi RNA binding protein 1 |
| chr2_+_200308943 | 0.92 |
ENST00000619961.4
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr10_+_60778331 | 0.86 |
ENST00000519078.6
ENST00000316629.8 ENST00000395284.8 |
CDK1
|
cyclin dependent kinase 1 |
| chr11_+_134331874 | 0.84 |
ENST00000339772.9
ENST00000535456.7 |
GLB1L2
|
galactosidase beta 1 like 2 |
| chr12_-_53507482 | 0.81 |
ENST00000267017.4
ENST00000448979.4 |
NPFF
|
neuropeptide FF-amide peptide precursor |
| chr7_-_15561986 | 0.81 |
ENST00000342526.8
|
AGMO
|
alkylglycerol monooxygenase |
| chr10_+_60778490 | 0.80 |
ENST00000448257.6
ENST00000614696.4 |
CDK1
|
cyclin dependent kinase 1 |
| chr3_-_99850976 | 0.80 |
ENST00000487087.5
|
FILIP1L
|
filamin A interacting protein 1 like |
| chr12_-_70637405 | 0.76 |
ENST00000548122.2
ENST00000551525.5 ENST00000550358.5 ENST00000334414.11 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr10_-_96271553 | 0.73 |
ENST00000224337.10
|
BLNK
|
B cell linker |
| chr15_-_42156590 | 0.72 |
ENST00000397272.7
|
PLA2G4F
|
phospholipase A2 group IVF |
| chr17_+_79022908 | 0.70 |
ENST00000354124.7
ENST00000580454.5 |
C1QTNF1
|
C1q and TNF related 1 |
| chr2_-_48755569 | 0.68 |
ENST00000428232.2
ENST00000405626.5 |
LHCGR
|
luteinizing hormone/choriogonadotropin receptor |
| chr15_-_33068143 | 0.64 |
ENST00000558197.1
|
FMN1
|
formin 1 |
| chr7_-_151187212 | 0.64 |
ENST00000420175.3
ENST00000275838.5 |
ASB10
|
ankyrin repeat and SOCS box containing 10 |
| chr3_-_52535006 | 0.63 |
ENST00000307076.8
|
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chr13_-_75366973 | 0.60 |
ENST00000648194.1
|
TBC1D4
|
TBC1 domain family member 4 |
| chr4_+_105552611 | 0.59 |
ENST00000265154.6
ENST00000420470.3 |
ARHGEF38
|
Rho guanine nucleotide exchange factor 38 |
| chr10_-_96271508 | 0.58 |
ENST00000427367.6
ENST00000413476.6 ENST00000371176.6 |
BLNK
|
B cell linker |
| chr10_-_96271398 | 0.56 |
ENST00000495266.1
|
BLNK
|
B cell linker |
| chr1_+_81306096 | 0.56 |
ENST00000370721.5
ENST00000370727.5 ENST00000370725.5 ENST00000370723.5 ENST00000370728.5 ENST00000370730.5 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr8_-_67062120 | 0.52 |
ENST00000357849.9
|
COPS5
|
COP9 signalosome subunit 5 |
| chr14_+_73851809 | 0.49 |
ENST00000555661.6
ENST00000555228.5 |
PTGR2
|
prostaglandin reductase 2 |
| chr10_+_73110375 | 0.47 |
ENST00000617744.4
ENST00000544879.5 ENST00000537969.5 ENST00000357321.9 ENST00000349051.9 |
NUDT13
|
nudix hydrolase 13 |
| chr17_+_79022814 | 0.44 |
ENST00000578229.5
|
C1QTNF1
|
C1q and TNF related 1 |
| chr20_-_21397513 | 0.44 |
ENST00000351817.5
|
NKX2-4
|
NK2 homeobox 4 |
| chr14_+_73851908 | 0.44 |
ENST00000267568.8
|
PTGR2
|
prostaglandin reductase 2 |
| chr2_-_18560616 | 0.43 |
ENST00000381249.4
|
RDH14
|
retinol dehydrogenase 14 |
| chr2_-_31414694 | 0.40 |
ENST00000379416.4
|
XDH
|
xanthine dehydrogenase |
| chr15_+_41286011 | 0.40 |
ENST00000661438.1
|
ENSG00000285920.2
|
novel protein |
| chr3_+_9902619 | 0.37 |
ENST00000421412.5
|
IL17RE
|
interleukin 17 receptor E |
| chr3_-_108953762 | 0.37 |
ENST00000393963.7
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr7_-_151187772 | 0.32 |
ENST00000377867.7
|
ASB10
|
ankyrin repeat and SOCS box containing 10 |
| chr1_-_205994439 | 0.30 |
ENST00000617991.4
|
RAB7B
|
RAB7B, member RAS oncogene family |
| chr10_-_73096850 | 0.29 |
ENST00000307116.6
ENST00000373008.6 ENST00000394890.7 |
P4HA1
|
prolyl 4-hydroxylase subunit alpha 1 |
| chr3_-_108953870 | 0.27 |
ENST00000261047.8
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr1_-_160954801 | 0.26 |
ENST00000368029.4
|
ITLN2
|
intelectin 2 |
| chr10_+_73110494 | 0.25 |
ENST00000372997.3
|
NUDT13
|
nudix hydrolase 13 |
| chr1_-_46665849 | 0.25 |
ENST00000532925.5
ENST00000542495.5 |
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr10_-_73096974 | 0.22 |
ENST00000440381.5
ENST00000263556.3 |
P4HA1
|
prolyl 4-hydroxylase subunit alpha 1 |
| chr22_+_20116099 | 0.09 |
ENST00000430524.6
|
RANBP1
|
RAN binding protein 1 |
| chr14_-_21023318 | 0.09 |
ENST00000298684.9
ENST00000557169.5 ENST00000553563.5 |
NDRG2
|
NDRG family member 2 |
| chr1_+_1287109 | 0.09 |
ENST00000379099.3
|
SCNN1D
|
sodium channel epithelial 1 subunit delta |
| chr1_+_18631513 | 0.08 |
ENST00000400661.3
|
PAX7
|
paired box 7 |
| chr3_+_9902808 | 0.08 |
ENST00000383814.8
ENST00000454190.6 ENST00000454992.1 |
IL17RE
|
interleukin 17 receptor E |
| chr5_-_135895834 | 0.08 |
ENST00000274520.2
|
IL9
|
interleukin 9 |
| chr11_+_101914997 | 0.08 |
ENST00000263468.13
|
CEP126
|
centrosomal protein 126 |
| chr1_-_11803383 | 0.07 |
ENST00000641407.1
ENST00000376583.7 ENST00000423400.7 |
MTHFR
|
methylenetetrahydrofolate reductase |
| chr1_-_182672882 | 0.06 |
ENST00000367557.8
ENST00000258302.8 |
RGS8
|
regulator of G protein signaling 8 |
| chr7_+_144070313 | 0.04 |
ENST00000641441.1
|
OR2A25
|
olfactory receptor family 2 subfamily A member 25 |
| chr16_+_2205708 | 0.01 |
ENST00000397124.5
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog |
| chr17_-_62065248 | 0.00 |
ENST00000397786.7
|
MED13
|
mediator complex subunit 13 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DMC1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.5 | 4.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.4 | 1.2 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.4 | 1.1 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.3 | 1.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.3 | 1.1 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 | 1.7 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.2 | 4.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 1.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 0.8 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.1 | 3.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.4 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 2.9 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.1 | 0.8 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 4.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.8 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.6 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.3 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.6 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.9 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.5 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.6 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.7 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 0.0 | 1.5 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.9 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.7 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0030849 | autosome(GO:0030849) |
| 0.4 | 1.7 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.4 | 1.2 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.4 | 5.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 0.5 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 2.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 4.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.6 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.4 | 1.6 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.4 | 1.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 1.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 0.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 4.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 1.1 | GO:0019959 | interleukin-8 binding(GO:0019959) tumor necrosis factor binding(GO:0043120) |
| 0.1 | 0.8 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.9 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 0.6 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 2.9 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 3.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.8 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.6 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 1.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.4 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.8 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 1.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 4.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.8 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.9 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.6 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.7 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.9 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |