Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for DRGX_PROP1
Z-value: 0.96
Transcription factors associated with DRGX_PROP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
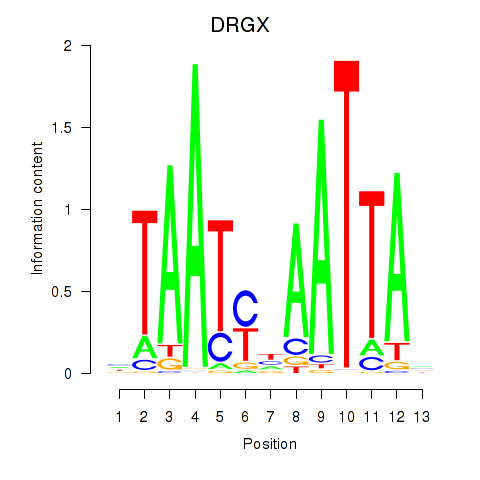
DRGX
|
ENSG00000165606.10 | dorsal root ganglia homeobox |
|
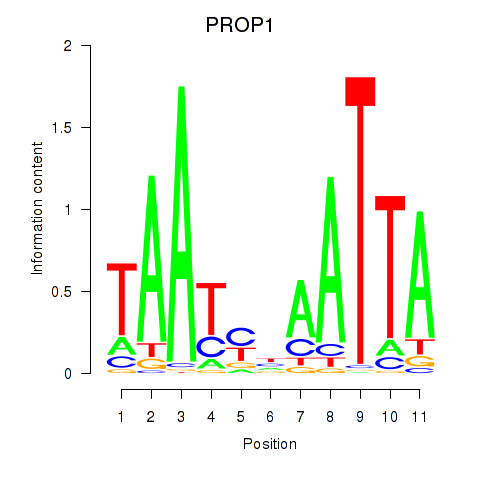
PROP1
|
ENSG00000175325.2 | PROP paired-like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DRGX | hg38_v1_chr10_-_49396074_49396098 | 0.09 | 6.1e-01 | Click! |
| PROP1 | hg38_v1_chr5_-_177996242_177996242 | 0.05 | 8.0e-01 | Click! |
Activity profile of DRGX_PROP1 motif
Sorted Z-values of DRGX_PROP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_68106109 | 5.80 |
ENST00000540630.5
ENST00000354393.6 |
MYPN
|
myopalladin |
| chr15_+_48191648 | 4.43 |
ENST00000646012.1
ENST00000561127.5 ENST00000647546.1 ENST00000559641.5 ENST00000417307.3 |
SLC12A1
CTXN2
|
solute carrier family 12 member 1 cortexin 2 |
| chr4_+_154563003 | 4.31 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr10_-_60141004 | 2.56 |
ENST00000355288.6
|
ANK3
|
ankyrin 3 |
| chr4_-_175812746 | 2.52 |
ENST00000393658.6
|
GPM6A
|
glycoprotein M6A |
| chr1_-_151175966 | 2.40 |
ENST00000441701.1
ENST00000295314.9 |
TMOD4
|
tropomodulin 4 |
| chr8_+_66126896 | 2.36 |
ENST00000353317.9
|
TRIM55
|
tripartite motif containing 55 |
| chr5_+_137867868 | 2.17 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chrX_-_55182442 | 2.03 |
ENST00000545075.3
|
MTRNR2L10
|
MT-RNR2 like 10 |
| chr2_-_182427014 | 1.95 |
ENST00000409365.5
ENST00000351439.9 |
PDE1A
|
phosphodiesterase 1A |
| chr10_-_60140515 | 1.91 |
ENST00000486349.2
|
ANK3
|
ankyrin 3 |
| chr5_+_137867852 | 1.76 |
ENST00000421631.6
ENST00000239926.9 |
MYOT
|
myotilin |
| chr11_+_18412292 | 1.76 |
ENST00000541669.6
ENST00000280704.8 |
LDHC
|
lactate dehydrogenase C |
| chr6_-_136550407 | 1.73 |
ENST00000354570.8
|
MAP7
|
microtubule associated protein 7 |
| chr2_+_28778848 | 1.69 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1 catalytic subunit beta |
| chr4_-_103019634 | 1.66 |
ENST00000510559.1
ENST00000296422.12 ENST00000394789.7 |
SLC9B1
|
solute carrier family 9 member B1 |
| chr2_+_161416273 | 1.65 |
ENST00000389554.8
|
TBR1
|
T-box brain transcription factor 1 |
| chr18_+_616672 | 1.58 |
ENST00000338387.11
|
CLUL1
|
clusterin like 1 |
| chr13_-_46897021 | 1.58 |
ENST00000542664.4
ENST00000543956.5 |
HTR2A
|
5-hydroxytryptamine receptor 2A |
| chr14_+_23240346 | 1.54 |
ENST00000430154.6
|
RNF212B
|
ring finger protein 212B |
| chr6_+_54099565 | 1.52 |
ENST00000511678.5
|
MLIP
|
muscular LMNA interacting protein |
| chr1_-_198540674 | 1.51 |
ENST00000489986.1
ENST00000367382.6 |
ATP6V1G3
|
ATPase H+ transporting V1 subunit G3 |
| chr18_+_616711 | 1.47 |
ENST00000579494.1
|
CLUL1
|
clusterin like 1 |
| chr7_-_83162899 | 1.41 |
ENST00000423517.6
|
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr7_+_123601836 | 1.37 |
ENST00000434204.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr7_+_123601815 | 1.36 |
ENST00000451215.6
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr6_+_54099538 | 1.34 |
ENST00000447836.6
|
MLIP
|
muscular LMNA interacting protein |
| chr17_+_70075215 | 1.31 |
ENST00000283936.5
ENST00000615244.4 ENST00000392671.6 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr2_+_44275457 | 1.28 |
ENST00000611973.4
ENST00000409387.5 |
SLC3A1
|
solute carrier family 3 member 1 |
| chr2_+_44275491 | 1.28 |
ENST00000410056.7
ENST00000409741.5 ENST00000409229.7 |
SLC3A1
|
solute carrier family 3 member 1 |
| chr2_+_233691607 | 1.27 |
ENST00000373424.5
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase family 1 member A6 |
| chr3_-_165196689 | 1.25 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chr1_+_42380772 | 1.25 |
ENST00000431473.4
ENST00000410070.6 |
RIMKLA
|
ribosomal modification protein rimK like family member A |
| chr9_-_123184233 | 1.24 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein |
| chr4_+_94974984 | 1.22 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr7_-_83162857 | 1.19 |
ENST00000333891.14
|
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr7_+_123601859 | 1.19 |
ENST00000437535.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr11_+_108116688 | 1.17 |
ENST00000672284.1
|
ACAT1
|
acetyl-CoA acetyltransferase 1 |
| chr7_-_117872209 | 1.15 |
ENST00000454375.5
|
CTTNBP2
|
cortactin binding protein 2 |
| chr2_+_44275473 | 1.15 |
ENST00000260649.11
|
SLC3A1
|
solute carrier family 3 member 1 |
| chr14_+_75069577 | 1.14 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger C2HC-type containing 1C |
| chr12_-_46825949 | 1.13 |
ENST00000547477.5
ENST00000447411.5 ENST00000266579.9 |
SLC38A4
|
solute carrier family 38 member 4 |
| chr11_-_26567087 | 1.12 |
ENST00000436318.6
ENST00000281268.12 |
MUC15
|
mucin 15, cell surface associated |
| chr3_-_9249623 | 1.11 |
ENST00000383836.8
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr13_-_101416441 | 1.10 |
ENST00000675332.1
ENST00000676315.1 ENST00000251127.11 |
NALCN
|
sodium leak channel, non-selective |
| chr5_+_90640718 | 1.09 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr17_+_73232400 | 1.08 |
ENST00000535032.7
ENST00000577615.5 ENST00000585109.5 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr12_+_75391078 | 1.07 |
ENST00000550916.6
ENST00000378692.7 ENST00000320460.8 ENST00000547164.1 |
GLIPR1L2
|
GLIPR1 like 2 |
| chr2_+_165294031 | 1.03 |
ENST00000283256.10
|
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr12_-_119804298 | 1.02 |
ENST00000678652.1
ENST00000678494.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr11_-_26572254 | 1.01 |
ENST00000529533.6
|
MUC15
|
mucin 15, cell surface associated |
| chr12_-_75390890 | 0.99 |
ENST00000552497.5
ENST00000551829.5 ENST00000436898.5 |
CAPS2
|
calcyphosine 2 |
| chr3_-_165196369 | 0.99 |
ENST00000475390.2
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chr4_-_99435134 | 0.98 |
ENST00000476959.5
ENST00000482593.5 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr5_-_11589019 | 0.97 |
ENST00000511377.5
|
CTNND2
|
catenin delta 2 |
| chrX_+_108045050 | 0.97 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr5_-_138139382 | 0.96 |
ENST00000265191.4
|
NME5
|
NME/NM23 family member 5 |
| chr17_+_73232637 | 0.94 |
ENST00000268942.12
ENST00000426147.6 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr17_+_70075317 | 0.93 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr12_-_119804472 | 0.93 |
ENST00000678087.1
ENST00000677993.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr13_+_53028806 | 0.90 |
ENST00000219022.3
|
OLFM4
|
olfactomedin 4 |
| chr12_-_9999176 | 0.89 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chrX_-_15314543 | 0.88 |
ENST00000344384.8
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr2_-_55334529 | 0.86 |
ENST00000645860.1
ENST00000642563.1 ENST00000647396.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chrX_+_108044967 | 0.85 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr1_+_50103903 | 0.85 |
ENST00000371827.5
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr18_+_58196736 | 0.83 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr4_+_48483324 | 0.82 |
ENST00000273861.5
|
SLC10A4
|
solute carrier family 10 member 4 |
| chr3_-_33645433 | 0.82 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr17_+_73232704 | 0.80 |
ENST00000582391.1
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chr14_-_75069478 | 0.79 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1 |
| chr1_-_21050952 | 0.77 |
ENST00000264211.12
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma 3 |
| chr2_-_55269038 | 0.77 |
ENST00000417363.5
ENST00000412530.1 ENST00000366137.6 ENST00000420637.5 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr11_-_26572130 | 0.76 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr17_+_73232601 | 0.76 |
ENST00000359042.6
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chr12_+_28452493 | 0.76 |
ENST00000542801.5
|
CCDC91
|
coiled-coil domain containing 91 |
| chr6_+_87407965 | 0.74 |
ENST00000369562.9
|
CFAP206
|
cilia and flagella associated protein 206 |
| chr5_-_11588842 | 0.74 |
ENST00000503622.5
|
CTNND2
|
catenin delta 2 |
| chr16_-_67483541 | 0.73 |
ENST00000290953.3
|
AGRP
|
agouti related neuropeptide |
| chr4_+_87650277 | 0.73 |
ENST00000339673.11
ENST00000282479.8 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr21_-_14210884 | 0.72 |
ENST00000679868.1
ENST00000400211.3 ENST00000680801.1 ENST00000536861.6 ENST00000614229.5 |
LIPI
|
lipase I |
| chr11_-_26572102 | 0.71 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated |
| chr12_-_44921842 | 0.71 |
ENST00000552993.5
|
NELL2
|
neural EGFL like 2 |
| chr5_-_135954962 | 0.70 |
ENST00000522943.5
ENST00000514447.2 ENST00000274507.6 |
LECT2
|
leukocyte cell derived chemotaxin 2 |
| chr7_-_123199960 | 0.70 |
ENST00000194130.7
|
SLC13A1
|
solute carrier family 13 member 1 |
| chr3_-_197573323 | 0.70 |
ENST00000358186.6
ENST00000431056.5 |
BDH1
|
3-hydroxybutyrate dehydrogenase 1 |
| chr21_-_14210948 | 0.69 |
ENST00000681601.1
|
LIPI
|
lipase I |
| chr6_+_34236865 | 0.68 |
ENST00000674029.1
ENST00000447654.5 ENST00000347617.10 ENST00000401473.7 ENST00000311487.9 |
HMGA1
|
high mobility group AT-hook 1 |
| chr2_+_100821025 | 0.68 |
ENST00000427413.5
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr8_-_17895403 | 0.67 |
ENST00000381840.5
ENST00000398054.5 |
FGL1
|
fibrinogen like 1 |
| chr9_-_27005659 | 0.67 |
ENST00000380055.6
|
LRRC19
|
leucine rich repeat containing 19 |
| chr6_-_41071825 | 0.66 |
ENST00000468811.5
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr11_-_13496018 | 0.65 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr9_+_85941529 | 0.64 |
ENST00000376040.2
|
NAA35
|
N-alpha-acetyltransferase 35, NatC auxiliary subunit |
| chr6_-_134317905 | 0.62 |
ENST00000461976.2
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr3_-_12545499 | 0.62 |
ENST00000564146.4
|
MKRN2OS
|
MKRN2 opposite strand |
| chr7_+_130207847 | 0.61 |
ENST00000297819.4
|
SSMEM1
|
serine rich single-pass membrane protein 1 |
| chr6_+_3258888 | 0.61 |
ENST00000380305.4
|
PSMG4
|
proteasome assembly chaperone 4 |
| chr4_+_94995919 | 0.61 |
ENST00000509540.6
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr4_+_94995796 | 0.59 |
ENST00000506363.5
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr5_+_170504005 | 0.59 |
ENST00000328939.9
ENST00000390656.8 |
KCNIP1
|
potassium voltage-gated channel interacting protein 1 |
| chr2_-_178794944 | 0.57 |
ENST00000436599.1
|
TTN
|
titin |
| chr8_-_17895487 | 0.57 |
ENST00000427924.5
ENST00000381841.4 |
FGL1
|
fibrinogen like 1 |
| chr9_+_12693327 | 0.57 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
| chr5_+_122160168 | 0.56 |
ENST00000509403.6
ENST00000514637.1 |
ENSG00000250803.6
|
novel zinc finger protein |
| chr10_+_104275126 | 0.55 |
ENST00000369707.2
|
GSTO2
|
glutathione S-transferase omega 2 |
| chr22_+_25219633 | 0.55 |
ENST00000398215.3
|
CRYBB2
|
crystallin beta B2 |
| chr4_+_93203949 | 0.54 |
ENST00000512631.1
|
GRID2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr4_-_175891691 | 0.54 |
ENST00000507540.1
|
GPM6A
|
glycoprotein M6A |
| chr12_-_118359639 | 0.53 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr12_+_102957666 | 0.53 |
ENST00000266744.4
|
ASCL1
|
achaete-scute family bHLH transcription factor 1 |
| chr7_-_120858066 | 0.52 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12 |
| chr19_-_57974527 | 0.50 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr5_-_55173173 | 0.50 |
ENST00000296733.5
ENST00000322374.10 ENST00000381375.7 |
CDC20B
|
cell division cycle 20B |
| chr3_+_42936859 | 0.50 |
ENST00000446977.2
ENST00000418176.1 |
ENSG00000273291.5
KRBOX1
|
novel protein KRAB box domain containing 1 |
| chr5_-_161546708 | 0.50 |
ENST00000393959.6
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr4_-_99435336 | 0.49 |
ENST00000437033.7
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr17_+_46624036 | 0.49 |
ENST00000575068.5
|
NSF
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr11_-_58731936 | 0.49 |
ENST00000344743.8
|
GLYAT
|
glycine-N-acyltransferase |
| chr5_+_173918216 | 0.48 |
ENST00000519467.1
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr1_+_84181630 | 0.48 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr2_+_184598520 | 0.47 |
ENST00000302277.7
|
ZNF804A
|
zinc finger protein 804A |
| chr10_+_35175586 | 0.46 |
ENST00000494479.5
ENST00000463314.5 ENST00000342105.7 ENST00000495301.1 |
CREM
|
cAMP responsive element modulator |
| chr4_-_99435396 | 0.46 |
ENST00000209665.8
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr2_-_58241259 | 0.46 |
ENST00000481670.2
ENST00000403676.5 ENST00000427708.6 ENST00000403295.7 ENST00000233741.9 ENST00000446381.5 ENST00000417361.1 ENST00000402135.7 ENST00000449070.5 |
FANCL
|
FA complementation group L |
| chr5_-_56116946 | 0.45 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chrX_+_120604199 | 0.45 |
ENST00000371315.3
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr19_+_984377 | 0.45 |
ENST00000591997.2
|
WDR18
|
WD repeat domain 18 |
| chr15_+_45252228 | 0.45 |
ENST00000560438.5
ENST00000347644.8 |
SLC28A2
|
solute carrier family 28 member 2 |
| chr5_+_173918186 | 0.44 |
ENST00000657000.1
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr12_-_118359105 | 0.44 |
ENST00000541186.5
ENST00000539872.5 |
TAOK3
|
TAO kinase 3 |
| chr6_-_134318097 | 0.44 |
ENST00000367858.10
ENST00000533224.1 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chrX_+_18707638 | 0.43 |
ENST00000472826.5
ENST00000496075.2 |
PPEF1
|
protein phosphatase with EF-hand domain 1 |
| chr12_+_100503352 | 0.43 |
ENST00000551379.5
ENST00000188403.7 |
NR1H4
|
nuclear receptor subfamily 1 group H member 4 |
| chr1_+_170145959 | 0.42 |
ENST00000439373.3
|
METTL11B
|
methyltransferase like 11B |
| chr2_+_86907953 | 0.42 |
ENST00000409776.6
|
RGPD1
|
RANBP2 like and GRIP domain containing 1 |
| chr4_-_52020332 | 0.42 |
ENST00000682860.1
|
LRRC66
|
leucine rich repeat containing 66 |
| chr5_-_11588796 | 0.42 |
ENST00000513598.5
|
CTNND2
|
catenin delta 2 |
| chr17_-_9791586 | 0.40 |
ENST00000571134.2
|
DHRS7C
|
dehydrogenase/reductase 7C |
| chr6_+_63576864 | 0.40 |
ENST00000627002.1
|
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr12_+_100503416 | 0.39 |
ENST00000551184.1
|
NR1H4
|
nuclear receptor subfamily 1 group H member 4 |
| chr5_-_127073467 | 0.39 |
ENST00000607731.5
ENST00000509733.7 ENST00000296662.10 ENST00000535381.6 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr6_+_3258914 | 0.39 |
ENST00000438998.7
ENST00000419065.6 ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome assembly chaperone 4 |
| chr12_+_19205294 | 0.39 |
ENST00000424268.5
|
PLEKHA5
|
pleckstrin homology domain containing A5 |
| chr11_+_89924064 | 0.38 |
ENST00000623787.3
|
TRIM49D2
|
tripartite motif containing 49D2 |
| chr1_+_77918128 | 0.38 |
ENST00000342754.5
|
NEXN
|
nexilin F-actin binding protein |
| chr1_+_248508073 | 0.37 |
ENST00000641804.1
|
OR2G6
|
olfactory receptor family 2 subfamily G member 6 |
| chr14_-_81427390 | 0.37 |
ENST00000555447.5
|
STON2
|
stonin 2 |
| chr9_-_34397800 | 0.37 |
ENST00000297623.7
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chrX_+_77910656 | 0.36 |
ENST00000343533.9
ENST00000341514.11 ENST00000645454.1 ENST00000642651.1 ENST00000644362.1 |
ATP7A
PGK1
|
ATPase copper transporting alpha phosphoglycerate kinase 1 |
| chr3_-_33645253 | 0.36 |
ENST00000333778.10
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr8_-_56211257 | 0.35 |
ENST00000316981.8
ENST00000423799.6 ENST00000429357.2 |
PLAG1
|
PLAG1 zinc finger |
| chr1_+_244051275 | 0.35 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr19_-_7021431 | 0.34 |
ENST00000636986.2
ENST00000637800.1 |
MBD3L2B
|
methyl-CpG binding domain protein 3 like 2B |
| chr17_-_40867200 | 0.34 |
ENST00000647902.1
ENST00000251643.5 |
KRT12
|
keratin 12 |
| chr12_-_118359166 | 0.33 |
ENST00000542902.5
|
TAOK3
|
TAO kinase 3 |
| chr19_-_56840661 | 0.33 |
ENST00000649735.1
ENST00000593695.5 ENST00000599577.5 ENST00000594389.1 ENST00000648694.1 ENST00000326441.15 ENST00000649680.1 ENST00000649876.1 ENST00000650632.1 ENST00000650102.1 ENST00000647621.1 ENST00000598410.5 ENST00000649233.1 ENST00000593711.6 ENST00000629319.2 ENST00000599935.5 |
PEG3
ZIM2
|
paternally expressed 3 zinc finger imprinted 2 |
| chr9_+_74615582 | 0.33 |
ENST00000396204.2
|
RORB
|
RAR related orphan receptor B |
| chr11_+_49028823 | 0.33 |
ENST00000332682.9
|
TRIM49B
|
tripartite motif containing 49B |
| chr9_+_121567057 | 0.33 |
ENST00000394340.7
ENST00000436835.5 ENST00000259371.6 |
DAB2IP
|
DAB2 interacting protein |
| chr3_+_2892199 | 0.33 |
ENST00000397459.6
|
CNTN4
|
contactin 4 |
| chr7_-_143408848 | 0.32 |
ENST00000275815.4
|
EPHA1
|
EPH receptor A1 |
| chr8_-_73259502 | 0.32 |
ENST00000624510.3
ENST00000613105.4 ENST00000625134.1 |
C8orf89
|
chromosome 8 open reading frame 89 |
| chr5_-_160852200 | 0.32 |
ENST00000327245.10
|
ATP10B
|
ATPase phospholipid transporting 10B (putative) |
| chr3_+_88338451 | 0.32 |
ENST00000637986.2
|
CSNKA2IP
|
casein kinase 2 subunit alpha' interacting protein |
| chr5_+_140827950 | 0.31 |
ENST00000378126.4
ENST00000529310.6 ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr13_-_45201027 | 0.31 |
ENST00000379108.2
|
KCTD4
|
potassium channel tetramerization domain containing 4 |
| chr8_+_12104389 | 0.31 |
ENST00000400085.7
|
ZNF705D
|
zinc finger protein 705D |
| chr11_-_16397521 | 0.31 |
ENST00000533411.5
|
SOX6
|
SRY-box transcription factor 6 |
| chr22_-_28711931 | 0.31 |
ENST00000434810.5
ENST00000456369.5 |
CHEK2
|
checkpoint kinase 2 |
| chr2_+_149038685 | 0.31 |
ENST00000409642.8
ENST00000409876.5 |
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr11_-_11353241 | 0.31 |
ENST00000528848.3
|
CSNK2A3
|
casein kinase 2 alpha 3 |
| chrX_+_147943245 | 0.30 |
ENST00000463120.2
|
FMR1
|
FMRP translational regulator 1 |
| chr4_+_112647059 | 0.30 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein 7, transcriptional regulator |
| chr9_-_135639540 | 0.30 |
ENST00000371763.6
ENST00000613244.1 |
GLT6D1
|
glycosyltransferase 6 domain containing 1 |
| chr4_+_75724569 | 0.30 |
ENST00000514213.7
ENST00000264904.8 |
USO1
|
USO1 vesicle transport factor |
| chr4_+_73481737 | 0.29 |
ENST00000226355.5
|
AFM
|
afamin |
| chr3_+_149474688 | 0.29 |
ENST00000305354.5
ENST00000465758.1 |
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr6_+_55327469 | 0.29 |
ENST00000340465.2
|
GFRAL
|
GDNF family receptor alpha like |
| chr7_+_112480853 | 0.29 |
ENST00000439068.6
ENST00000312849.4 |
LSMEM1
|
leucine rich single-pass membrane protein 1 |
| chr5_-_175961324 | 0.29 |
ENST00000432305.6
ENST00000505969.1 |
THOC3
|
THO complex 3 |
| chr11_-_57712205 | 0.28 |
ENST00000337672.9
ENST00000431606.4 |
MED19
|
mediator complex subunit 19 |
| chr3_+_111978996 | 0.28 |
ENST00000273359.8
ENST00000494817.1 |
ABHD10
|
abhydrolase domain containing 10, depalmitoylase |
| chr4_+_87832917 | 0.28 |
ENST00000395102.8
ENST00000497649.6 ENST00000540395.1 ENST00000560249.5 ENST00000511670.5 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr4_-_46124046 | 0.28 |
ENST00000295452.5
|
GABRG1
|
gamma-aminobutyric acid type A receptor subunit gamma1 |
| chr16_-_69339493 | 0.28 |
ENST00000562595.5
ENST00000615447.1 ENST00000306875.10 ENST00000562081.2 |
COG8
|
component of oligomeric golgi complex 8 |
| chr1_-_165445088 | 0.27 |
ENST00000359842.10
|
RXRG
|
retinoid X receptor gamma |
| chr5_+_142770367 | 0.27 |
ENST00000645722.2
ENST00000274498.9 |
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr12_+_71667783 | 0.27 |
ENST00000551238.1
|
THAP2
|
THAP domain containing 2 |
| chr11_-_58731968 | 0.26 |
ENST00000278400.3
|
GLYAT
|
glycine-N-acyltransferase |
| chr11_-_84166998 | 0.26 |
ENST00000398299.1
|
DLG2
|
discs large MAGUK scaffold protein 2 |
| chr17_-_40703744 | 0.26 |
ENST00000264651.3
|
KRT24
|
keratin 24 |
| chr14_-_80959484 | 0.26 |
ENST00000555529.5
ENST00000556042.5 ENST00000556981.5 |
CEP128
|
centrosomal protein 128 |
| chr11_-_40294089 | 0.26 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr9_+_65700287 | 0.26 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr11_+_22672577 | 0.26 |
ENST00000534801.5
|
GAS2
|
growth arrest specific 2 |
| chr4_-_183322426 | 0.25 |
ENST00000541814.1
|
CLDN24
|
claudin 24 |
| chr3_-_191282383 | 0.25 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr12_+_14419136 | 0.25 |
ENST00000545769.1
ENST00000428217.6 ENST00000396279.2 ENST00000542514.5 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr1_+_12857086 | 0.25 |
ENST00000240189.2
|
PRAMEF2
|
PRAME family member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DRGX_PROP1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.6 | 4.5 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.5 | 1.9 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.4 | 3.7 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.4 | 1.8 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.3 | 1.6 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.3 | 4.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 2.4 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.3 | 1.2 | GO:0072229 | isoleucine catabolic process(GO:0006550) acetyl-CoA catabolic process(GO:0046356) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.3 | 6.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 2.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 1.7 | GO:0021764 | amygdala development(GO:0021764) |
| 0.2 | 0.8 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.2 | 0.6 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.2 | 0.4 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.2 | 0.5 | GO:0021529 | noradrenergic neuron development(GO:0003358) spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.7 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.9 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.8 | GO:1903314 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.1 | 0.7 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.7 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.4 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.1 | 1.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.3 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 1.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 1.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.2 | GO:0042309 | octopamine biosynthetic process(GO:0006589) homoiothermy(GO:0042309) octopamine metabolic process(GO:0046333) |
| 0.1 | 0.2 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 1.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 2.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.3 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.2 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.2 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.1 | 1.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 1.0 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.1 | 0.8 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.0 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.3 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.1 | 1.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 0.5 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.5 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 0.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.2 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 0.7 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.8 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 1.0 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.4 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 3.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.6 | GO:0090487 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.0 | 0.3 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 1.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.7 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.7 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 3.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0002434 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) negative regulation of eosinophil activation(GO:1902567) positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 2.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.7 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 2.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 5.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.2 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.6 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.8 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.0 | 0.1 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 1.5 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 1.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 2.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.4 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 2.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.7 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.9 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 1.7 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.4 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.5 | GO:0010842 | retina layer formation(GO:0010842) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 0.4 | 5.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 0.7 | GO:0001534 | radial spoke(GO:0001534) |
| 0.2 | 2.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 4.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.7 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.6 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 1.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 3.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.3 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.1 | 0.3 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 2.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 1.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 10.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 1.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.1 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) translation initiation ternary complex(GO:0044207) |
| 0.0 | 0.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 3.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 3.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 2.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 2.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.7 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 2.6 | GO:0005795 | Golgi stack(GO:0005795) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.5 | 1.9 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.4 | 3.7 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.3 | 1.7 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.3 | 2.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.3 | 1.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.2 | 1.2 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.2 | 0.7 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 2.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 1.8 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 6.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 1.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.6 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.8 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.1 | 0.8 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.6 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.4 | GO:0004618 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 2.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.4 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.1 | 0.4 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 1.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.6 | GO:0050610 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.1 | 2.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.7 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.1 | 0.2 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 2.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.8 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.6 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 2.9 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 4.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.7 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 1.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.9 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 1.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.1 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.8 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 1.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.3 | GO:0034046 | RNA strand annealing activity(GO:0033592) poly(G) binding(GO:0034046) |
| 0.0 | 0.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.0 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 4.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.8 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.7 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 3.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.5 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) transferrin receptor binding(GO:1990459) |
| 0.0 | 2.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.1 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 3.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 3.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 4.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.7 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 2.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 4.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.8 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 4.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.7 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |