Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for DUXA
Z-value: 1.26
Transcription factors associated with DUXA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DUXA
|
ENSG00000258873.3 | double homeobox A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DUXA | hg38_v1_chr19_-_57167485_57167485 | -0.38 | 3.1e-02 | Click! |
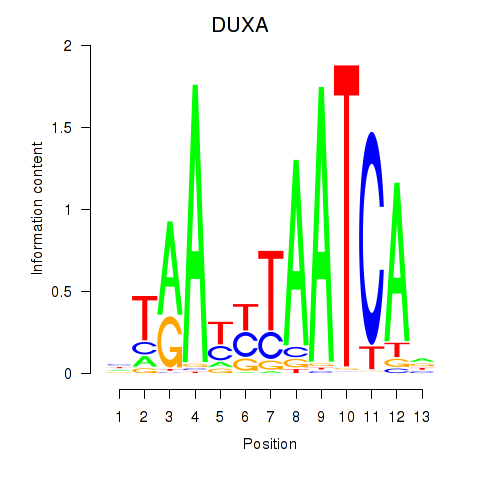
Activity profile of DUXA motif
Sorted Z-values of DUXA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_6483728 | 3.52 |
ENST00000675459.1
ENST00000551752.5 |
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr2_-_174764436 | 3.33 |
ENST00000409323.1
ENST00000261007.9 ENST00000348749.9 ENST00000672640.1 |
CHRNA1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr16_+_6483813 | 2.30 |
ENST00000675653.1
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr10_+_68106109 | 2.28 |
ENST00000540630.5
ENST00000354393.6 |
MYPN
|
myopalladin |
| chr6_+_54018910 | 2.12 |
ENST00000514921.5
ENST00000274897.9 ENST00000370877.6 |
MLIP
|
muscular LMNA interacting protein |
| chr3_-_196515315 | 2.07 |
ENST00000397537.3
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr10_-_67665642 | 1.99 |
ENST00000682945.1
ENST00000330298.6 ENST00000682758.1 |
CTNNA3
|
catenin alpha 3 |
| chr10_-_29522527 | 1.99 |
ENST00000632315.1
|
SVIL
|
supervillin |
| chr15_-_42457556 | 1.88 |
ENST00000565948.1
|
ZNF106
|
zinc finger protein 106 |
| chr5_+_142770367 | 1.84 |
ENST00000645722.2
ENST00000274498.9 |
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr3_+_160225409 | 1.73 |
ENST00000326474.5
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr8_-_106770235 | 1.69 |
ENST00000311955.4
|
ABRA
|
actin binding Rho activating protein |
| chr2_-_174764407 | 1.55 |
ENST00000409219.5
ENST00000409542.5 |
CHRNA1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr1_+_77918128 | 1.51 |
ENST00000342754.5
|
NEXN
|
nexilin F-actin binding protein |
| chr5_+_68239815 | 1.48 |
ENST00000520675.1
|
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr4_-_46124046 | 1.42 |
ENST00000295452.5
|
GABRG1
|
gamma-aminobutyric acid type A receptor subunit gamma1 |
| chr2_-_223602284 | 1.42 |
ENST00000421386.1
ENST00000305409.3 ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr6_+_54018992 | 1.41 |
ENST00000509997.5
|
MLIP
|
muscular LMNA interacting protein |
| chr5_+_174045673 | 1.41 |
ENST00000303177.8
ENST00000519867.5 |
NSG2
|
neuronal vesicle trafficking associated 2 |
| chr10_-_29634964 | 1.38 |
ENST00000375398.6
ENST00000355867.8 |
SVIL
|
supervillin |
| chr8_-_7056729 | 1.36 |
ENST00000330590.4
|
DEFA5
|
defensin alpha 5 |
| chr5_+_98773651 | 1.36 |
ENST00000513185.3
|
RGMB
|
repulsive guidance molecule BMP co-receptor b |
| chr5_-_79514127 | 1.32 |
ENST00000334082.11
|
HOMER1
|
homer scaffold protein 1 |
| chr18_-_77127935 | 1.29 |
ENST00000581878.5
|
MBP
|
myelin basic protein |
| chr3_-_39280021 | 1.29 |
ENST00000399220.3
|
CX3CR1
|
C-X3-C motif chemokine receptor 1 |
| chr16_+_6483379 | 1.24 |
ENST00000552089.5
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr7_+_80646436 | 1.24 |
ENST00000419819.2
|
CD36
|
CD36 molecule |
| chr1_-_13285154 | 1.22 |
ENST00000357367.6
ENST00000614831.1 |
PRAMEF8
|
PRAME family member 8 |
| chrX_+_155216452 | 1.14 |
ENST00000286428.7
|
VBP1
|
VHL binding protein 1 |
| chr10_-_60141004 | 1.12 |
ENST00000355288.6
|
ANK3
|
ankyrin 3 |
| chr19_-_50968125 | 1.10 |
ENST00000594641.1
|
KLK6
|
kallikrein related peptidase 6 |
| chr6_+_12717660 | 1.09 |
ENST00000674637.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr11_+_49028823 | 1.08 |
ENST00000332682.9
|
TRIM49B
|
tripartite motif containing 49B |
| chr11_-_133956957 | 1.07 |
ENST00000533871.8
|
IGSF9B
|
immunoglobulin superfamily member 9B |
| chr11_-_89807220 | 1.07 |
ENST00000532501.2
|
TRIM49
|
tripartite motif containing 49 |
| chr22_+_31122923 | 1.02 |
ENST00000620191.4
ENST00000412277.6 ENST00000412985.5 ENST00000331075.10 ENST00000420017.5 ENST00000400294.6 ENST00000405300.5 ENST00000404390.7 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr2_-_40453438 | 1.02 |
ENST00000455476.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr19_-_3061403 | 1.01 |
ENST00000586839.1
|
TLE5
|
TLE family member 5, transcriptional modulator |
| chr17_-_59081162 | 1.01 |
ENST00000581468.1
|
TRIM37
|
tripartite motif containing 37 |
| chr2_+_54115437 | 1.00 |
ENST00000303536.8
ENST00000394666.7 |
ACYP2
|
acylphosphatase 2 |
| chr1_+_12857086 | 0.99 |
ENST00000240189.2
|
PRAMEF2
|
PRAME family member 2 |
| chr1_-_42335869 | 0.98 |
ENST00000372573.5
|
FOXJ3
|
forkhead box J3 |
| chr2_+_148875214 | 0.97 |
ENST00000435030.6
ENST00000677891.1 ENST00000677843.1 ENST00000678056.1 ENST00000677280.1 |
KIF5C
|
kinesin family member 5C |
| chr18_-_3219849 | 0.95 |
ENST00000261606.11
|
MYOM1
|
myomesin 1 |
| chr4_-_175907143 | 0.93 |
ENST00000513365.1
ENST00000513667.5 ENST00000503563.1 |
GPM6A
|
glycoprotein M6A |
| chr1_+_12916610 | 0.93 |
ENST00000616979.4
|
PRAMEF7
|
PRAME family member 7 |
| chr18_-_3219961 | 0.92 |
ENST00000356443.9
|
MYOM1
|
myomesin 1 |
| chr5_+_36606355 | 0.92 |
ENST00000681909.1
ENST00000513903.5 ENST00000681795.1 ENST00000680125.1 ENST00000612708.5 ENST00000680232.1 ENST00000681776.1 ENST00000681926.1 ENST00000679958.1 ENST00000265113.9 ENST00000504121.5 ENST00000512374.1 ENST00000613445.5 ENST00000679983.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr19_-_50968775 | 0.91 |
ENST00000391808.5
|
KLK6
|
kallikrein related peptidase 6 |
| chr12_-_44921842 | 0.91 |
ENST00000552993.5
|
NELL2
|
neural EGFL like 2 |
| chr21_+_33025927 | 0.88 |
ENST00000430860.1
ENST00000382357.4 ENST00000333337.3 |
OLIG2
|
oligodendrocyte transcription factor 2 |
| chr16_-_21303036 | 0.87 |
ENST00000219599.8
ENST00000576703.5 |
CRYM
|
crystallin mu |
| chr4_+_48483324 | 0.87 |
ENST00000273861.5
|
SLC10A4
|
solute carrier family 10 member 4 |
| chr20_-_10420737 | 0.87 |
ENST00000649912.1
|
ENSG00000285723.1
|
novel protein |
| chr4_-_6555609 | 0.85 |
ENST00000507294.1
|
PPP2R2C
|
protein phosphatase 2 regulatory subunit Bgamma |
| chr12_-_10909562 | 0.82 |
ENST00000390677.2
|
TAS2R13
|
taste 2 receptor member 13 |
| chr1_+_13070853 | 0.81 |
ENST00000619661.2
|
PRAMEF25
|
PRAME family member 25 |
| chr1_+_12773738 | 0.80 |
ENST00000357726.5
|
PRAMEF12
|
PRAME family member 12 |
| chr5_-_17655843 | 0.80 |
ENST00000598383.3
|
H3Y1
|
H3.Y histone 1 |
| chr11_-_89920428 | 0.80 |
ENST00000605881.5
|
TRIM49D1
|
tripartite motif containing 49D1 |
| chr4_+_75724569 | 0.80 |
ENST00000514213.7
ENST00000264904.8 |
USO1
|
USO1 vesicle transport factor |
| chr5_+_17604177 | 0.79 |
ENST00000510838.2
|
TAF11L11
|
TATA-box binding protein associated factor 11 like 11 |
| chr14_-_80231052 | 0.79 |
ENST00000557010.5
|
DIO2
|
iodothyronine deiodinase 2 |
| chr1_+_13068677 | 0.78 |
ENST00000614839.4
|
PRAMEF25
|
PRAME family member 25 |
| chr16_-_57186014 | 0.78 |
ENST00000566584.5
ENST00000566481.5 ENST00000566077.5 ENST00000564108.5 ENST00000309137.13 ENST00000565458.5 ENST00000566681.1 ENST00000567439.5 |
PSME3IP1
|
proteasome activator subunit 3 interacting protein 1 |
| chrX_+_86714623 | 0.76 |
ENST00000484479.1
|
DACH2
|
dachshund family transcription factor 2 |
| chr7_-_14902743 | 0.76 |
ENST00000402815.6
|
DGKB
|
diacylglycerol kinase beta |
| chr4_-_141212877 | 0.75 |
ENST00000420921.6
|
RNF150
|
ring finger protein 150 |
| chr7_-_14902842 | 0.74 |
ENST00000407950.5
|
DGKB
|
diacylglycerol kinase beta |
| chr16_-_57185808 | 0.73 |
ENST00000562406.5
ENST00000568671.5 ENST00000567044.5 |
PSME3IP1
|
proteasome activator subunit 3 interacting protein 1 |
| chr6_-_70957029 | 0.71 |
ENST00000230053.11
|
B3GAT2
|
beta-1,3-glucuronyltransferase 2 |
| chr3_+_108602776 | 0.71 |
ENST00000497905.5
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr11_+_28108248 | 0.69 |
ENST00000406787.7
ENST00000403099.5 ENST00000407364.8 |
METTL15
|
methyltransferase like 15 |
| chr19_+_52429181 | 0.69 |
ENST00000301085.8
|
ZNF534
|
zinc finger protein 534 |
| chr17_+_68515399 | 0.69 |
ENST00000588188.6
|
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chr1_+_248095184 | 0.68 |
ENST00000358120.3
ENST00000641893.1 |
OR2L13
|
olfactory receptor family 2 subfamily L member 13 |
| chr9_-_104599413 | 0.68 |
ENST00000374779.3
|
OR13C5
|
olfactory receptor family 13 subfamily C member 5 |
| chr1_-_12898270 | 0.68 |
ENST00000235347.4
|
PRAMEF10
|
PRAME family member 10 |
| chr18_-_36798482 | 0.68 |
ENST00000590258.2
|
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr7_-_14903319 | 0.68 |
ENST00000403951.6
|
DGKB
|
diacylglycerol kinase beta |
| chr11_-_4288083 | 0.67 |
ENST00000638166.1
|
SSU72P4
|
SSU72 pseudogene 4 |
| chr4_-_185810894 | 0.65 |
ENST00000448662.6
ENST00000439049.5 ENST00000420158.5 ENST00000319471.13 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_-_41459592 | 0.65 |
ENST00000528697.6
ENST00000530763.5 |
LRRC4C
|
leucine rich repeat containing 4C |
| chr11_-_133956754 | 0.65 |
ENST00000321016.12
|
IGSF9B
|
immunoglobulin superfamily member 9B |
| chr2_+_86106217 | 0.64 |
ENST00000409783.6
ENST00000254630.12 ENST00000627371.1 |
PTCD3
|
pentatricopeptide repeat domain 3 |
| chr2_-_68871382 | 0.64 |
ENST00000295379.2
|
BMP10
|
bone morphogenetic protein 10 |
| chr12_-_89656051 | 0.64 |
ENST00000261173.6
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr5_+_69217721 | 0.63 |
ENST00000256441.5
|
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr19_-_7021431 | 0.63 |
ENST00000636986.2
ENST00000637800.1 |
MBD3L2B
|
methyl-CpG binding domain protein 3 like 2B |
| chr15_-_26629083 | 0.63 |
ENST00000400188.7
|
GABRB3
|
gamma-aminobutyric acid type A receptor subunit beta3 |
| chr2_+_165572329 | 0.62 |
ENST00000342316.8
|
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr11_-_48983826 | 0.62 |
ENST00000649162.1
|
TRIM51GP
|
tripartite motif-containing 51G, pseudogene |
| chr1_-_165445088 | 0.61 |
ENST00000359842.10
|
RXRG
|
retinoid X receptor gamma |
| chr8_-_17002327 | 0.61 |
ENST00000180166.6
|
FGF20
|
fibroblast growth factor 20 |
| chr6_+_72366730 | 0.60 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr21_+_42199686 | 0.60 |
ENST00000398457.6
|
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr1_-_220046432 | 0.60 |
ENST00000609181.5
ENST00000366923.8 |
EPRS1
|
glutamyl-prolyl-tRNA synthetase 1 |
| chr2_-_88861258 | 0.60 |
ENST00000390240.2
|
IGKJ3
|
immunoglobulin kappa joining 3 |
| chr19_+_7030578 | 0.60 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3 like 5 |
| chr4_-_69787955 | 0.59 |
ENST00000512870.1
|
SULT1B1
|
sulfotransferase family 1B member 1 |
| chr11_+_89924064 | 0.59 |
ENST00000623787.3
|
TRIM49D2
|
tripartite motif containing 49D2 |
| chr11_-_123654581 | 0.58 |
ENST00000392770.6
ENST00000530277.5 ENST00000299333.8 |
SCN3B
|
sodium voltage-gated channel beta subunit 3 |
| chr15_+_62561361 | 0.58 |
ENST00000561311.5
|
TLN2
|
talin 2 |
| chr16_-_57946602 | 0.57 |
ENST00000564654.1
|
CNGB1
|
cyclic nucleotide gated channel subunit beta 1 |
| chr19_+_7049321 | 0.57 |
ENST00000381393.3
|
MBD3L2
|
methyl-CpG binding domain protein 3 like 2 |
| chr5_+_31532277 | 0.57 |
ENST00000507818.6
ENST00000325366.14 |
C5orf22
|
chromosome 5 open reading frame 22 |
| chr1_+_74198310 | 0.55 |
ENST00000472069.1
|
FPGT
|
fucose-1-phosphate guanylyltransferase |
| chr19_-_6393205 | 0.55 |
ENST00000595047.5
|
GTF2F1
|
general transcription factor IIF subunit 1 |
| chr11_+_99021066 | 0.54 |
ENST00000527185.5
ENST00000528682.5 |
CNTN5
|
contactin 5 |
| chr7_+_142462882 | 0.54 |
ENST00000454561.2
|
TRBV5-4
|
T cell receptor beta variable 5-4 |
| chr5_-_111976925 | 0.54 |
ENST00000395634.7
|
NREP
|
neuronal regeneration related protein |
| chr5_+_17610496 | 0.53 |
ENST00000503184.2
|
TAF11L12
|
TATA-box binding protein associated factor 11 like 12 |
| chr5_-_88731827 | 0.53 |
ENST00000627170.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr11_-_123654939 | 0.53 |
ENST00000657191.1
|
SCN3B
|
sodium voltage-gated channel beta subunit 3 |
| chr4_-_75724386 | 0.53 |
ENST00000677606.1
ENST00000678798.1 ENST00000677970.1 ENST00000677620.1 ENST00000679281.1 ENST00000677333.1 ENST00000676470.1 ENST00000499709.3 ENST00000511868.6 ENST00000678971.1 ENST00000677265.1 ENST00000677952.1 ENST00000678122.1 ENST00000678100.1 ENST00000678062.1 ENST00000676666.1 |
G3BP2
|
G3BP stress granule assembly factor 2 |
| chr6_-_55579178 | 0.53 |
ENST00000308161.8
ENST00000398661.6 ENST00000274901.9 |
HMGCLL1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase like 1 |
| chr1_-_12947580 | 0.52 |
ENST00000376189.5
|
PRAMEF6
|
PRAME family member 6 |
| chr11_+_5351508 | 0.52 |
ENST00000380219.1
|
OR51B6
|
olfactory receptor family 51 subfamily B member 6 |
| chr11_+_55262152 | 0.52 |
ENST00000417545.5
|
TRIM48
|
tripartite motif containing 48 |
| chr3_-_165196689 | 0.52 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chr3_+_45886501 | 0.52 |
ENST00000395963.2
|
CCR9
|
C-C motif chemokine receptor 9 |
| chr4_-_185813121 | 0.52 |
ENST00000456060.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr15_+_70936487 | 0.52 |
ENST00000558456.5
ENST00000560158.6 ENST00000558808.5 ENST00000559806.5 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr8_-_12134099 | 0.51 |
ENST00000530447.5
|
USP17L7
|
ubiquitin specific peptidase 17 like family member 7 |
| chr2_+_1484663 | 0.51 |
ENST00000446278.5
ENST00000469607.3 |
TPO
|
thyroid peroxidase |
| chr11_-_4242640 | 0.51 |
ENST00000640805.1
|
SSU72P2
|
SSU72 pseudogene 2 |
| chr11_-_13496018 | 0.50 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr1_-_13201409 | 0.50 |
ENST00000625019.3
|
PRAMEF13
|
PRAME family member 13 |
| chr1_+_13303539 | 0.50 |
ENST00000437300.2
|
PRAMEF33
|
PRAME family member 33 |
| chr12_+_120302316 | 0.50 |
ENST00000536460.1
ENST00000202967.4 |
SIRT4
|
sirtuin 4 |
| chr1_-_12886201 | 0.50 |
ENST00000235349.6
|
PRAMEF4
|
PRAME family member 4 |
| chr16_-_21211566 | 0.49 |
ENST00000574091.6
|
ZP2
|
zona pellucida glycoprotein 2 |
| chr10_-_49188312 | 0.49 |
ENST00000453436.5
ENST00000474718.5 |
TMEM273
|
transmembrane protein 273 |
| chr1_-_12831410 | 0.48 |
ENST00000619922.1
|
PRAMEF11
|
PRAME family member 11 |
| chr11_-_4339244 | 0.48 |
ENST00000524542.2
|
SSU72P7
|
SSU72 pseudogene 7 |
| chr17_+_68525795 | 0.48 |
ENST00000592800.5
|
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chr19_-_7058640 | 0.48 |
ENST00000333843.8
|
MBD3L3
|
methyl-CpG binding domain protein 3 like 3 |
| chr8_+_91101832 | 0.48 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr19_-_2090132 | 0.47 |
ENST00000591326.5
|
MOB3A
|
MOB kinase activator 3A |
| chr12_-_89656093 | 0.47 |
ENST00000359142.7
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr4_+_107989880 | 0.47 |
ENST00000309522.8
ENST00000403312.6 ENST00000638559.1 ENST00000682373.1 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr12_+_12725897 | 0.46 |
ENST00000326765.10
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr1_-_12945416 | 0.46 |
ENST00000415464.6
|
PRAMEF6
|
PRAME family member 6 |
| chr1_-_207052980 | 0.46 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr2_+_197516039 | 0.46 |
ENST00000448447.6
ENST00000323303.9 ENST00000409360.1 |
MOB4
|
MOB family member 4, phocein |
| chr18_-_72865680 | 0.46 |
ENST00000397929.5
|
NETO1
|
neuropilin and tolloid like 1 |
| chr11_+_99020940 | 0.45 |
ENST00000524871.6
|
CNTN5
|
contactin 5 |
| chr8_-_81695045 | 0.45 |
ENST00000518568.3
|
SLC10A5
|
solute carrier family 10 member 5 |
| chr17_-_4263847 | 0.45 |
ENST00000570535.5
ENST00000574367.5 ENST00000341657.9 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr9_+_136982993 | 0.45 |
ENST00000408973.3
|
LCNL1
|
lipocalin like 1 |
| chr4_+_653171 | 0.45 |
ENST00000488061.5
ENST00000429163.6 |
PDE6B
|
phosphodiesterase 6B |
| chr13_+_57147488 | 0.44 |
ENST00000377930.1
|
PRR20B
|
proline rich 20B |
| chr12_-_6635938 | 0.44 |
ENST00000329858.9
|
LPAR5
|
lysophosphatidic acid receptor 5 |
| chr7_+_124476371 | 0.44 |
ENST00000473520.1
|
SSU72P8
|
SSU72 pseudogene 8 |
| chr9_-_6008469 | 0.44 |
ENST00000399933.8
|
KIAA2026
|
KIAA2026 |
| chr6_-_55579160 | 0.44 |
ENST00000370850.6
|
HMGCLL1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase like 1 |
| chr2_-_196809352 | 0.44 |
ENST00000342506.4
|
C2orf66
|
chromosome 2 open reading frame 66 |
| chr11_-_55607645 | 0.44 |
ENST00000641580.1
|
OR4C11
|
olfactory receptor family 4 subfamily C member 11 |
| chr16_-_57186053 | 0.44 |
ENST00000565760.5
ENST00000570184.1 ENST00000562324.5 |
PSME3IP1
|
proteasome activator subunit 3 interacting protein 1 |
| chr12_-_109996216 | 0.43 |
ENST00000551209.5
ENST00000550186.5 |
GIT2
|
GIT ArfGAP 2 |
| chr11_+_4233288 | 0.43 |
ENST00000639584.1
|
SSU72P5
|
SSU72 pseudogene 5 |
| chr9_-_28670285 | 0.43 |
ENST00000379992.6
ENST00000308675.5 ENST00000613945.3 |
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr1_+_74198230 | 0.43 |
ENST00000557284.7
ENST00000370899.7 ENST00000370895.5 ENST00000534632.5 ENST00000370893.1 ENST00000467578.7 ENST00000370894.9 ENST00000482102.2 ENST00000370898.9 ENST00000534056.5 |
FPGT-TNNI3K
FPGT
|
FPGT-TNNI3K readthrough fucose-1-phosphate guanylyltransferase |
| chr10_+_70093626 | 0.43 |
ENST00000678195.1
|
MACROH2A2
|
macroH2A.2 histone |
| chrX_+_150699208 | 0.43 |
ENST00000436701.5
ENST00000438018.5 |
MTMR1
|
myotubularin related protein 1 |
| chr15_-_92809798 | 0.42 |
ENST00000557398.2
|
FAM174B
|
family with sequence similarity 174 member B |
| chr11_-_117817500 | 0.42 |
ENST00000525836.1
|
DSCAML1
|
DS cell adhesion molecule like 1 |
| chr5_-_31532039 | 0.42 |
ENST00000511367.6
ENST00000344624.8 ENST00000513349.5 |
DROSHA
|
drosha ribonuclease III |
| chr1_-_165445220 | 0.42 |
ENST00000619224.1
|
RXRG
|
retinoid X receptor gamma |
| chr2_-_212124901 | 0.41 |
ENST00000402597.6
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4 |
| chr12_-_119877300 | 0.41 |
ENST00000392521.7
|
CIT
|
citron rho-interacting serine/threonine kinase |
| chr7_-_126533850 | 0.41 |
ENST00000444921.3
|
GRM8
|
glutamate metabotropic receptor 8 |
| chr2_-_88861920 | 0.41 |
ENST00000390242.2
|
IGKJ1
|
immunoglobulin kappa joining 1 |
| chr19_-_6393131 | 0.41 |
ENST00000394456.10
|
GTF2F1
|
general transcription factor IIF subunit 1 |
| chr14_+_22147988 | 0.41 |
ENST00000390457.2
|
TRAV27
|
T cell receptor alpha variable 27 |
| chr11_+_123902167 | 0.40 |
ENST00000641687.1
|
OR8D4
|
olfactory receptor family 8 subfamily D member 4 |
| chr2_+_108607140 | 0.40 |
ENST00000410093.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chrX_+_53094133 | 0.40 |
ENST00000604062.6
|
KANTR
|
KDM5C adjacent transcript |
| chr2_-_197198034 | 0.40 |
ENST00000328737.6
|
ANKRD44
|
ankyrin repeat domain 44 |
| chr15_+_43330646 | 0.40 |
ENST00000562188.7
ENST00000428046.7 ENST00000389651.8 |
ADAL
|
adenosine deaminase like |
| chr6_+_121437378 | 0.40 |
ENST00000650427.1
ENST00000647564.1 |
GJA1
|
gap junction protein alpha 1 |
| chrX_+_65488735 | 0.40 |
ENST00000338957.4
|
ZC3H12B
|
zinc finger CCCH-type containing 12B |
| chr8_-_12138845 | 0.39 |
ENST00000333796.4
|
USP17L2
|
ubiquitin specific peptidase 17 like family member 2 |
| chr1_-_248559516 | 0.39 |
ENST00000328570.6
|
OR2T29
|
olfactory receptor family 2 subfamily T member 29 |
| chr16_-_70289480 | 0.39 |
ENST00000261772.13
ENST00000675953.1 ENST00000675691.1 ENST00000675133.1 ENST00000674512.1 ENST00000675045.1 ENST00000675853.1 ENST00000565361.3 ENST00000674963.1 ENST00000674691.1 |
AARS1
|
alanyl-tRNA synthetase 1 |
| chr17_-_41397600 | 0.39 |
ENST00000251645.3
|
KRT31
|
keratin 31 |
| chr16_-_29995601 | 0.39 |
ENST00000279392.8
ENST00000564026.1 |
HIRIP3
|
HIRA interacting protein 3 |
| chr6_-_73225465 | 0.39 |
ENST00000370388.4
|
KHDC1L
|
KH domain containing 1 like |
| chr7_+_142535763 | 0.38 |
ENST00000614171.1
|
TRBV13
|
T cell receptor beta variable 13 |
| chr12_-_118359639 | 0.38 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr1_+_13254212 | 0.38 |
ENST00000622421.2
|
PRAMEF5
|
PRAME family member 5 |
| chr11_+_118359572 | 0.37 |
ENST00000252108.8
ENST00000431736.6 |
UBE4A
|
ubiquitination factor E4A |
| chr11_+_55883297 | 0.37 |
ENST00000449290.6
|
TRIM51
|
tripartite motif-containing 51 |
| chr11_+_24496988 | 0.37 |
ENST00000336930.11
|
LUZP2
|
leucine zipper protein 2 |
| chr19_-_14674829 | 0.37 |
ENST00000443157.6
ENST00000253673.6 |
ADGRE3
|
adhesion G protein-coupled receptor E3 |
| chr20_-_35411963 | 0.37 |
ENST00000349714.9
ENST00000438533.5 ENST00000359226.6 ENST00000374384.6 ENST00000374385.10 ENST00000424405.5 ENST00000397554.5 ENST00000374380.6 |
UQCC1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr8_-_53251412 | 0.36 |
ENST00000520287.5
|
OPRK1
|
opioid receptor kappa 1 |
| chr9_+_105662457 | 0.36 |
ENST00000334077.6
|
TAL2
|
TAL bHLH transcription factor 2 |
| chr4_-_75724362 | 0.36 |
ENST00000677583.1
|
G3BP2
|
G3BP stress granule assembly factor 2 |
| chr1_+_50103903 | 0.36 |
ENST00000371827.5
|
ELAVL4
|
ELAV like RNA binding protein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DUXA
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.9 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.3 | 1.0 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.3 | 1.3 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.3 | 0.9 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.3 | 2.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.3 | 1.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 1.3 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.2 | 0.6 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.2 | 0.4 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.2 | 2.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.2 | 0.5 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.2 | 3.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 0.5 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.1 | 0.5 | GO:1903217 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.1 | 0.9 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.4 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 1.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 1.1 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.1 | 0.4 | GO:1903937 | response to acrylamide(GO:1903937) |
| 0.1 | 0.4 | GO:0060003 | copper ion export(GO:0060003) |
| 0.1 | 0.4 | GO:0014873 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 | 0.5 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.9 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.3 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.1 | 0.5 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 2.0 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.3 | GO:0048561 | establishment of organ orientation(GO:0048561) |
| 0.1 | 1.2 | GO:0072564 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.3 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.1 | 0.3 | GO:0070145 | mitochondrial asparaginyl-tRNA aminoacylation(GO:0070145) |
| 0.1 | 1.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 0.3 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.1 | 0.6 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.1 | 0.5 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.4 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.1 | 2.3 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 1.0 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.3 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.1 | 0.3 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 4.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.4 | GO:2000984 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.1 | 0.4 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.5 | GO:0007343 | egg activation(GO:0007343) negative regulation of fertilization(GO:0060467) |
| 0.1 | 0.7 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 6.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 2.9 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 0.4 | GO:0032439 | endosome localization(GO:0032439) positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 0.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 1.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.6 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 1.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 2.0 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 | 0.4 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.1 | 1.5 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.1 | 0.2 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.1 | 0.2 | GO:1902960 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.1 | 0.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 1.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:1990172 | CVT pathway(GO:0032258) G-protein coupled receptor catabolic process(GO:1990172) |
| 0.0 | 0.5 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.9 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.2 | GO:0061015 | snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.6 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.2 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.7 | GO:2000628 | production of siRNA involved in RNA interference(GO:0030422) regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.1 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.4 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 1.0 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0002035 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.3 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.0 | 1.0 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 1.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.4 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 1.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.2 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 1.8 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 1.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.3 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 1.0 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.2 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.6 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.4 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 1.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.2 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.0 | 6.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 1.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 1.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 1.1 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 1.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0042746 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.6 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 1.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.2 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0034505 | tooth mineralization(GO:0034505) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.3 | 1.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.2 | 0.7 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.2 | 1.5 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.2 | 3.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 4.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.6 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 1.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 1.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.3 | GO:0099569 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 0.1 | 2.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.7 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 1.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 1.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 2.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) translation initiation ternary complex(GO:0044207) |
| 0.0 | 1.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 1.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 2.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 7.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 1.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0060342 | photoreceptor inner segment membrane(GO:0060342) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 7.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 2.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.7 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.2 | 1.0 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.2 | 1.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 0.9 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.2 | 1.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 2.0 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 1.0 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 4.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.6 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.7 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.5 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 1.0 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.1 | 0.7 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.4 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 0.5 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 1.0 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 4.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 1.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.5 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 1.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.3 | GO:0050560 | aspartate-tRNA ligase activity(GO:0004815) aspartate-tRNA(Asn) ligase activity(GO:0050560) |
| 0.1 | 0.7 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.7 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.4 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.1 | 2.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.5 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.3 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 1.0 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.1 | 0.6 | GO:0005223 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 1.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 0.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 1.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 2.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.6 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 1.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 1.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.8 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 1.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.4 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 1.0 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.6 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.3 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.5 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 1.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 1.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.4 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.5 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 5.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.9 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 5.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.4 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 6.8 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 2.8 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 2.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 2.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 3.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.7 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 1.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 2.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 2.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.1 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 4.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |