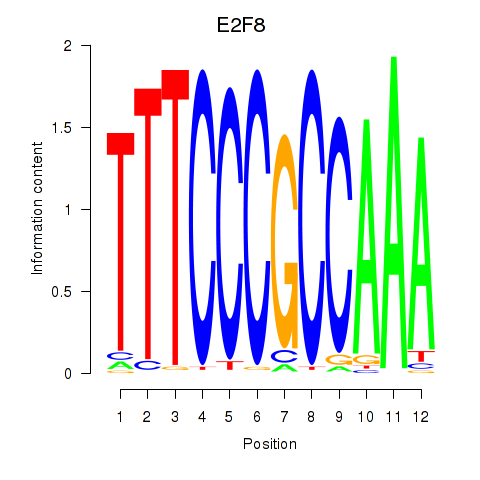
|
chr5_+_40909490
Show fit
|
1.94 |
ENST00000313164.10
|
C7
|
complement C7
|
|
chr19_+_40597168
Show fit
|
1.71 |
ENST00000308370.11
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr2_+_79025678
Show fit
|
1.69 |
ENST00000393897.6
|
REG3G
|
regenerating family member 3 gamma
|
|
chr2_+_79025696
Show fit
|
1.69 |
ENST00000272324.10
|
REG3G
|
regenerating family member 3 gamma
|
|
chr15_-_88913362
Show fit
|
1.53 |
ENST00000558029.5
ENST00000268150.13
ENST00000542878.5
ENST00000268151.11
ENST00000566497.5
|
MFGE8
|
milk fat globule EGF and factor V/VIII domain containing
|
|
chr8_+_17577179
Show fit
|
1.52 |
ENST00000251630.11
|
PDGFRL
|
platelet derived growth factor receptor like
|
|
chr17_-_4560564
Show fit
|
1.44 |
ENST00000574584.1
ENST00000381550.8
ENST00000301395.7
|
GGT6
|
gamma-glutamyltransferase 6
|
|
chr19_+_40601342
Show fit
|
1.36 |
ENST00000396819.8
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr4_+_15339818
Show fit
|
1.34 |
ENST00000397700.6
ENST00000295297.4
|
C1QTNF7
|
C1q and TNF related 7
|
|
chr12_+_20368495
Show fit
|
1.33 |
ENST00000359062.4
|
PDE3A
|
phosphodiesterase 3A
|
|
chr4_+_54229261
Show fit
|
1.32 |
ENST00000508170.5
ENST00000512143.1
ENST00000257290.10
|
PDGFRA
|
platelet derived growth factor receptor alpha
|
|
chr6_+_135181268
Show fit
|
1.31 |
ENST00000341911.10
ENST00000442647.7
ENST00000618728.4
ENST00000316528.12
ENST00000616088.4
|
MYB
|
MYB proto-oncogene, transcription factor
|
|
chr16_-_31135699
Show fit
|
1.21 |
ENST00000317508.11
ENST00000568261.5
ENST00000567797.1
|
PRSS8
|
serine protease 8
|
|
chr2_+_79025709
Show fit
|
1.15 |
ENST00000409471.1
|
REG3G
|
regenerating family member 3 gamma
|
|
chr2_-_130198433
Show fit
|
1.11 |
ENST00000312988.9
|
TUBA3E
|
tubulin alpha 3e
|
|
chr14_+_101561351
Show fit
|
1.05 |
ENST00000510508.4
|
DIO3
|
iodothyronine deiodinase 3
|
|
chr2_-_113235443
Show fit
|
1.04 |
ENST00000465084.1
|
PAX8
|
paired box 8
|
|
chr2_+_131476112
Show fit
|
1.03 |
ENST00000321253.7
|
TUBA3D
|
tubulin alpha 3d
|
|
chr19_-_46788586
Show fit
|
1.02 |
ENST00000542575.6
|
SLC1A5
|
solute carrier family 1 member 5
|
|
chr13_-_19181773
Show fit
|
0.98 |
ENST00000618094.1
ENST00000400113.8
|
TUBA3C
|
tubulin alpha 3c
|
|
chr14_+_24368020
Show fit
|
0.96 |
ENST00000554050.5
ENST00000554903.1
ENST00000250373.9
ENST00000554779.1
ENST00000553708.5
|
NFATC4
|
nuclear factor of activated T cells 4
|
|
chr11_+_9573633
Show fit
|
0.92 |
ENST00000450114.7
|
WEE1
|
WEE1 G2 checkpoint kinase
|
|
chr1_-_16980607
Show fit
|
0.92 |
ENST00000375535.4
|
MFAP2
|
microfibril associated protein 2
|
|
chr4_+_15427998
Show fit
|
0.88 |
ENST00000444304.3
|
C1QTNF7
|
C1q and TNF related 7
|
|
chr4_+_54229843
Show fit
|
0.86 |
ENST00000503856.5
|
PDGFRA
|
platelet derived growth factor receptor alpha
|
|
chr6_+_135181361
Show fit
|
0.85 |
ENST00000527615.5
ENST00000420123.6
ENST00000525369.5
ENST00000528774.5
ENST00000533624.5
ENST00000534044.5
ENST00000534121.5
|
MYB
|
MYB proto-oncogene, transcription factor
|
|
chr6_-_57221402
Show fit
|
0.84 |
ENST00000317483.4
|
RAB23
|
RAB23, member RAS oncogene family
|
|
chr1_-_150720842
Show fit
|
0.82 |
ENST00000442853.5
ENST00000368995.8
ENST00000322343.11
ENST00000361824.7
|
HORMAD1
|
HORMA domain containing 1
|
|
chr19_-_49361475
Show fit
|
0.81 |
ENST00000598810.5
|
TEAD2
|
TEA domain transcription factor 2
|
|
chr19_-_49362376
Show fit
|
0.79 |
ENST00000601519.5
ENST00000593945.6
ENST00000539846.5
ENST00000596757.1
ENST00000311227.6
|
TEAD2
|
TEA domain transcription factor 2
|
|
chr2_-_197785313
Show fit
|
0.77 |
ENST00000392296.9
|
BOLL
|
boule homolog, RNA binding protein
|
|
chr2_-_27139395
Show fit
|
0.76 |
ENST00000432962.2
ENST00000335524.7
|
PRR30
|
proline rich 30
|
|
chr14_-_61281310
Show fit
|
0.75 |
ENST00000555868.2
|
TMEM30B
|
transmembrane protein 30B
|
|
chr12_-_8662619
Show fit
|
0.75 |
ENST00000544889.1
ENST00000543369.5
|
MFAP5
|
microfibril associated protein 5
|
|
chr17_+_41819201
Show fit
|
0.73 |
ENST00000455106.1
|
FKBP10
|
FKBP prolyl isomerase 10
|
|
chr17_-_15262537
Show fit
|
0.72 |
ENST00000395936.7
ENST00000675819.1
ENST00000674707.1
ENST00000675854.1
ENST00000426385.4
ENST00000395938.7
ENST00000612492.5
ENST00000675808.1
|
PMP22
|
peripheral myelin protein 22
|
|
chr17_-_76585808
Show fit
|
0.71 |
ENST00000225276.10
|
ST6GALNAC2
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 2
|
|
chr3_-_100114488
Show fit
|
0.70 |
ENST00000477258.2
ENST00000354552.7
ENST00000331335.9
ENST00000398326.2
|
FILIP1L
|
filamin A interacting protein 1 like
|
|
chr1_+_150272772
Show fit
|
0.70 |
ENST00000369098.3
ENST00000369099.8
|
C1orf54
|
chromosome 1 open reading frame 54
|
|
chr16_-_57536543
Show fit
|
0.69 |
ENST00000258214.3
|
CCDC102A
|
coiled-coil domain containing 102A
|
|
chr6_+_135181323
Show fit
|
0.68 |
ENST00000367814.8
|
MYB
|
MYB proto-oncogene, transcription factor
|
|
chr3_+_69936629
Show fit
|
0.66 |
ENST00000394348.2
ENST00000531774.1
|
MITF
|
melanocyte inducing transcription factor
|
|
chr19_+_49388243
Show fit
|
0.65 |
ENST00000447857.8
|
KASH5
|
KASH domain containing 5
|
|
chr11_-_107858561
Show fit
|
0.65 |
ENST00000375682.8
|
SLC35F2
|
solute carrier family 35 member F2
|
|
chr10_-_17617235
Show fit
|
0.64 |
ENST00000466335.1
|
HACD1
|
3-hydroxyacyl-CoA dehydratase 1
|
|
chr16_+_11345429
Show fit
|
0.64 |
ENST00000576027.1
ENST00000312499.6
ENST00000648619.1
|
RMI2
|
RecQ mediated genome instability 2
|
|
chr11_-_107858777
Show fit
|
0.63 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2
|
|
chr16_-_31135640
Show fit
|
0.61 |
ENST00000567531.5
|
PRSS8
|
serine protease 8
|
|
chr12_+_106774630
Show fit
|
0.60 |
ENST00000392839.6
ENST00000548914.5
ENST00000355478.6
ENST00000552619.1
ENST00000549643.5
ENST00000392837.9
|
RIC8B
|
RIC8 guanine nucleotide exchange factor B
|
|
chr14_-_23155302
Show fit
|
0.60 |
ENST00000529705.6
|
SLC7A8
|
solute carrier family 7 member 8
|
|
chr2_-_24123251
Show fit
|
0.60 |
ENST00000313213.5
ENST00000436622.1
|
PFN4
|
profilin family member 4
|
|
chr16_+_84842003
Show fit
|
0.59 |
ENST00000563066.5
|
CRISPLD2
|
cysteine rich secretory protein LCCL domain containing 2
|
|
chr2_+_12716893
Show fit
|
0.58 |
ENST00000381465.2
ENST00000155926.9
|
TRIB2
|
tribbles pseudokinase 2
|
|
chr3_-_51875597
Show fit
|
0.57 |
ENST00000446461.2
|
IQCF5
|
IQ motif containing F5
|
|
chr1_+_43172324
Show fit
|
0.57 |
ENST00000528956.5
ENST00000610710.4
ENST00000372492.9
ENST00000529956.5
|
CFAP57
|
cilia and flagella associated protein 57
|
|
chr22_+_19479826
Show fit
|
0.57 |
ENST00000437685.6
ENST00000263201.7
ENST00000404724.7
|
CDC45
|
cell division cycle 45
|
|
chr12_+_1629197
Show fit
|
0.56 |
ENST00000397196.7
|
WNT5B
|
Wnt family member 5B
|
|
chr6_-_2962097
Show fit
|
0.56 |
ENST00000380524.5
|
SERPINB6
|
serpin family B member 6
|
|
chr10_+_110644306
Show fit
|
0.56 |
ENST00000369519.4
|
RBM20
|
RNA binding motif protein 20
|
|
chr4_+_20253500
Show fit
|
0.56 |
ENST00000622093.4
ENST00000273739.9
|
SLIT2
|
slit guidance ligand 2
|
|
chrX_+_52751132
Show fit
|
0.55 |
ENST00000616191.4
ENST00000596480.6
ENST00000612490.1
|
SSX2B
|
SSX family member 2B
|
|
chr1_+_15756603
Show fit
|
0.55 |
ENST00000496928.6
ENST00000508310.5
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr12_-_8662703
Show fit
|
0.55 |
ENST00000535336.5
|
MFAP5
|
microfibril associated protein 5
|
|
chr4_-_173333672
Show fit
|
0.55 |
ENST00000438704.6
|
HMGB2
|
high mobility group box 2
|
|
chr18_+_75210789
Show fit
|
0.54 |
ENST00000580243.3
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr11_-_7706469
Show fit
|
0.54 |
ENST00000533663.6
|
OVCH2
|
ovochymase 2
|
|
chr11_-_114559866
Show fit
|
0.53 |
ENST00000534921.2
|
NXPE1
|
neurexophilin and PC-esterase domain family member 1
|
|
chr7_-_100827504
Show fit
|
0.52 |
ENST00000616502.4
ENST00000358173.8
|
EPHB4
|
EPH receptor B4
|
|
chr10_+_116324440
Show fit
|
0.52 |
ENST00000333254.4
|
CCDC172
|
coiled-coil domain containing 172
|
|
chr6_-_84227634
Show fit
|
0.52 |
ENST00000617909.1
ENST00000403245.8
|
CEP162
|
centrosomal protein 162
|
|
chr2_+_69013414
Show fit
|
0.52 |
ENST00000681816.1
ENST00000482235.2
|
ANTXR1
|
ANTXR cell adhesion molecule 1
|
|
chr17_+_39738317
Show fit
|
0.51 |
ENST00000394211.7
|
GRB7
|
growth factor receptor bound protein 7
|
|
chr18_+_75210755
Show fit
|
0.51 |
ENST00000322038.5
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr6_-_29005313
Show fit
|
0.50 |
ENST00000377179.4
|
ZNF311
|
zinc finger protein 311
|
|
chr1_-_150720829
Show fit
|
0.50 |
ENST00000368987.5
|
HORMAD1
|
HORMA domain containing 1
|
|
chr7_+_128739395
Show fit
|
0.50 |
ENST00000479257.5
|
CALU
|
calumenin
|
|
chrX_+_136532205
Show fit
|
0.50 |
ENST00000370634.8
|
VGLL1
|
vestigial like family member 1
|
|
chr19_+_6740878
Show fit
|
0.49 |
ENST00000596673.1
|
TRIP10
|
thyroid hormone receptor interactor 10
|
|
chr7_+_128739292
Show fit
|
0.49 |
ENST00000535011.6
ENST00000542996.6
ENST00000249364.9
ENST00000449187.6
|
CALU
|
calumenin
|
|
chr11_+_72227881
Show fit
|
0.49 |
ENST00000538751.5
ENST00000541756.5
|
INPPL1
|
inositol polyphosphate phosphatase like 1
|
|
chrX_-_10620534
Show fit
|
0.49 |
ENST00000317552.9
|
MID1
|
midline 1
|
|
chr11_-_14971179
Show fit
|
0.47 |
ENST00000486207.5
|
CALCA
|
calcitonin related polypeptide alpha
|
|
chrX_-_52707161
Show fit
|
0.47 |
ENST00000336777.9
ENST00000337502.5
|
SSX2
|
SSX family member 2
|
|
chr1_-_159923717
Show fit
|
0.46 |
ENST00000368096.5
|
TAGLN2
|
transgelin 2
|
|
chr15_+_40439721
Show fit
|
0.46 |
ENST00000561234.5
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr2_+_200378218
Show fit
|
0.46 |
ENST00000449647.5
|
SPATS2L
|
spermatogenesis associated serine rich 2 like
|
|
chr17_+_7281711
Show fit
|
0.46 |
ENST00000317370.13
ENST00000571308.5
|
SLC2A4
|
solute carrier family 2 member 4
|
|
chr2_+_237487239
Show fit
|
0.46 |
ENST00000338530.8
ENST00000264605.8
ENST00000409373.5
|
MLPH
|
melanophilin
|
|
chr3_+_197950509
Show fit
|
0.46 |
ENST00000442341.5
|
RPL35A
|
ribosomal protein L35a
|
|
chr9_-_35079923
Show fit
|
0.45 |
ENST00000378643.8
ENST00000448890.1
|
FANCG
|
FA complementation group G
|
|
chr17_+_50426242
Show fit
|
0.44 |
ENST00000502667.5
|
ACSF2
|
acyl-CoA synthetase family member 2
|
|
chr19_-_51342130
Show fit
|
0.44 |
ENST00000335624.5
|
VSIG10L
|
V-set and immunoglobulin domain containing 10 like
|
|
chrX_+_132023294
Show fit
|
0.43 |
ENST00000481105.5
ENST00000354719.10
ENST00000394334.7
ENST00000394335.6
|
STK26
|
serine/threonine kinase 26
|
|
chr6_-_165987954
Show fit
|
0.43 |
ENST00000672902.1
|
PDE10A
|
phosphodiesterase 10A
|
|
chr17_+_50426210
Show fit
|
0.43 |
ENST00000506582.5
ENST00000504392.5
ENST00000300441.9
ENST00000427954.6
|
ACSF2
|
acyl-CoA synthetase family member 2
|
|
chr1_+_244835616
Show fit
|
0.42 |
ENST00000366528.3
ENST00000411948.7
|
COX20
|
cytochrome c oxidase assembly factor COX20
|
|
chr19_+_52297157
Show fit
|
0.42 |
ENST00000595962.6
ENST00000598016.5
ENST00000334564.11
ENST00000490272.1
|
ZNF480
|
zinc finger protein 480
|
|
chr1_+_35269016
Show fit
|
0.42 |
ENST00000441447.1
|
ZMYM4
|
zinc finger MYM-type containing 4
|
|
chr5_+_134371561
Show fit
|
0.41 |
ENST00000265339.7
ENST00000506787.5
ENST00000507277.1
|
UBE2B
|
ubiquitin conjugating enzyme E2 B
|
|
chr17_-_38967393
Show fit
|
0.41 |
ENST00000378079.3
|
FBXO47
|
F-box protein 47
|
|
chr14_-_50668287
Show fit
|
0.41 |
ENST00000556735.1
ENST00000324679.5
|
SAV1
|
salvador family WW domain containing protein 1
|
|
chrX_+_24054931
Show fit
|
0.41 |
ENST00000253039.9
ENST00000423068.1
|
EIF2S3
|
eukaryotic translation initiation factor 2 subunit gamma
|
|
chr12_-_70609788
Show fit
|
0.41 |
ENST00000547715.1
ENST00000538708.5
ENST00000550857.5
ENST00000261266.9
|
PTPRB
|
protein tyrosine phosphatase receptor type B
|
|
chr12_-_102062079
Show fit
|
0.40 |
ENST00000545679.5
|
WASHC3
|
WASH complex subunit 3
|
|
chr12_+_48106094
Show fit
|
0.40 |
ENST00000546755.5
ENST00000549366.5
ENST00000642730.1
ENST00000552792.5
|
PFKM
|
phosphofructokinase, muscle
|
|
chr4_+_105552611
Show fit
|
0.40 |
ENST00000265154.6
ENST00000420470.3
|
ARHGEF38
|
Rho guanine nucleotide exchange factor 38
|
|
chr1_+_35268663
Show fit
|
0.40 |
ENST00000314607.11
|
ZMYM4
|
zinc finger MYM-type containing 4
|
|
chr3_-_136752361
Show fit
|
0.39 |
ENST00000480733.1
ENST00000629124.2
ENST00000383202.7
ENST00000236698.9
ENST00000434713.6
|
STAG1
|
stromal antigen 1
|
|
chr19_+_49361783
Show fit
|
0.39 |
ENST00000594268.1
|
DKKL1
|
dickkopf like acrosomal protein 1
|
|
chr2_+_127620000
Show fit
|
0.39 |
ENST00000409090.1
|
MYO7B
|
myosin VIIB
|
|
chr3_+_44712634
Show fit
|
0.39 |
ENST00000449836.5
ENST00000296091.8
ENST00000436624.7
ENST00000411443.1
|
ZNF502
|
zinc finger protein 502
|
|
chr6_-_84227596
Show fit
|
0.39 |
ENST00000257766.8
|
CEP162
|
centrosomal protein 162
|
|
chr2_-_55617584
Show fit
|
0.39 |
ENST00000616288.4
ENST00000611717.4
ENST00000616407.2
|
PPP4R3B
|
protein phosphatase 4 regulatory subunit 3B
|
|
chr5_-_180861248
Show fit
|
0.39 |
ENST00000502412.2
ENST00000512132.5
ENST00000506439.5
|
ZFP62
|
ZFP62 zinc finger protein
|
|
chr16_-_66801578
Show fit
|
0.38 |
ENST00000433154.6
ENST00000673664.1
ENST00000559050.1
ENST00000558713.6
|
TERB1
|
telomere repeat binding bouquet formation protein 1
|
|
chr11_+_125626229
Show fit
|
0.38 |
ENST00000532449.6
ENST00000534070.5
|
CHEK1
|
checkpoint kinase 1
|
|
chr11_-_66677748
Show fit
|
0.38 |
ENST00000525754.5
ENST00000531969.5
ENST00000524637.1
ENST00000531036.2
ENST00000310046.9
|
RBM4B
|
RNA binding motif protein 4B
|
|
chr12_-_102061946
Show fit
|
0.37 |
ENST00000240079.11
|
WASHC3
|
WASH complex subunit 3
|
|
chr11_-_16356538
Show fit
|
0.37 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6
|
|
chr7_-_137343688
Show fit
|
0.37 |
ENST00000348225.7
|
PTN
|
pleiotrophin
|
|
chr17_-_75289212
Show fit
|
0.37 |
ENST00000582778.1
ENST00000581988.5
ENST00000579207.5
ENST00000583332.5
ENST00000442286.6
ENST00000580151.5
ENST00000580994.5
ENST00000584438.1
ENST00000416858.7
ENST00000320362.7
|
SLC25A19
|
solute carrier family 25 member 19
|
|
chr7_-_137343752
Show fit
|
0.36 |
ENST00000393083.2
|
PTN
|
pleiotrophin
|
|
chr8_-_23404076
Show fit
|
0.36 |
ENST00000524168.1
ENST00000389131.8
ENST00000523833.2
ENST00000519243.1
|
LOXL2
|
lysyl oxidase like 2
|
|
chr16_-_69351778
Show fit
|
0.36 |
ENST00000288025.4
|
TMED6
|
transmembrane p24 trafficking protein 6
|
|
chr9_+_136796331
Show fit
|
0.36 |
ENST00000338005.7
|
CCDC183
|
coiled-coil domain containing 183
|
|
chr14_-_55191534
Show fit
|
0.36 |
ENST00000395425.6
ENST00000247191.7
|
DLGAP5
|
DLG associated protein 5
|
|
chr7_-_100428657
Show fit
|
0.36 |
ENST00000360951.8
ENST00000398027.6
ENST00000684423.1
ENST00000472716.1
|
ZCWPW1
|
zinc finger CW-type and PWWP domain containing 1
|
|
chr12_+_2959870
Show fit
|
0.36 |
ENST00000397122.6
|
TEAD4
|
TEA domain transcription factor 4
|
|
chr16_-_82011444
Show fit
|
0.35 |
ENST00000328945.7
ENST00000532128.5
|
SDR42E1
|
short chain dehydrogenase/reductase family 42E, member 1
|
|
chr14_-_24429665
Show fit
|
0.35 |
ENST00000267406.11
|
CBLN3
|
cerebellin 3 precursor
|
|
chrX_+_106201724
Show fit
|
0.35 |
ENST00000372552.1
|
PWWP3B
|
PWWP domain containing 3B
|
|
chr15_+_43826961
Show fit
|
0.34 |
ENST00000381246.6
ENST00000452115.1
ENST00000263795.11
|
WDR76
|
WD repeat domain 76
|
|
chr17_+_49132763
Show fit
|
0.33 |
ENST00000393354.7
|
B4GALNT2
|
beta-1,4-N-acetyl-galactosaminyltransferase 2
|
|
chr19_-_35228699
Show fit
|
0.33 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187 member B
|
|
chr2_+_73784189
Show fit
|
0.32 |
ENST00000409561.1
|
C2orf78
|
chromosome 2 open reading frame 78
|
|
chr14_+_63761836
Show fit
|
0.32 |
ENST00000674003.1
|
SYNE2
|
spectrin repeat containing nuclear envelope protein 2
|
|
chr6_-_165987914
Show fit
|
0.32 |
ENST00000672859.1
|
PDE10A
|
phosphodiesterase 10A
|
|
chr18_+_74315836
Show fit
|
0.32 |
ENST00000579455.2
|
C18orf63
|
chromosome 18 open reading frame 63
|
|
chr12_-_6689450
Show fit
|
0.32 |
ENST00000355772.8
ENST00000417772.7
ENST00000319770.7
ENST00000396801.7
|
ZNF384
|
zinc finger protein 384
|
|
chr10_+_35127162
Show fit
|
0.31 |
ENST00000354759.7
|
CREM
|
cAMP responsive element modulator
|
|
chr6_+_10585748
Show fit
|
0.31 |
ENST00000265012.5
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group)
|
|
chr10_-_102418748
Show fit
|
0.31 |
ENST00000020673.6
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr11_+_125625967
Show fit
|
0.31 |
ENST00000428830.6
ENST00000278916.8
ENST00000544373.5
ENST00000532669.5
ENST00000527013.6
ENST00000526937.5
ENST00000534685.5
|
CHEK1
|
checkpoint kinase 1
|
|
chr19_+_50384323
Show fit
|
0.31 |
ENST00000599857.7
ENST00000613923.6
ENST00000601098.6
ENST00000440232.7
ENST00000595904.6
ENST00000593887.1
|
POLD1
|
DNA polymerase delta 1, catalytic subunit
|
|
chr12_+_109900518
Show fit
|
0.31 |
ENST00000312777.9
ENST00000536408.2
|
TCHP
|
trichoplein keratin filament binding
|
|
chr13_+_37000774
Show fit
|
0.31 |
ENST00000389704.4
|
EXOSC8
|
exosome component 8
|
|
chr14_+_20684547
Show fit
|
0.31 |
ENST00000555835.3
ENST00000397995.2
ENST00000553909.1
|
RNASE4
ENSG00000259171.1
|
ribonuclease A family member 4
novel protein, ANG-RNASE4 readthrough
|
|
chr19_+_49388276
Show fit
|
0.30 |
ENST00000598730.5
ENST00000594905.5
ENST00000595828.5
ENST00000594043.5
|
KASH5
|
KASH domain containing 5
|
|
chr2_-_86563470
Show fit
|
0.30 |
ENST00000409225.2
|
CHMP3
|
charged multivesicular body protein 3
|
|
chr19_+_35641728
Show fit
|
0.30 |
ENST00000619399.4
ENST00000379026.6
ENST00000379023.8
ENST00000402764.6
ENST00000479824.5
|
ETV2
|
ETS variant transcription factor 2
|
|
chr1_+_159010002
Show fit
|
0.30 |
ENST00000359709.7
|
IFI16
|
interferon gamma inducible protein 16
|
|
chr6_+_32024278
Show fit
|
0.29 |
ENST00000647698.1
|
C4B
|
complement C4B (Chido blood group)
|
|
chr12_+_48105466
Show fit
|
0.29 |
ENST00000549003.5
ENST00000550924.6
|
PFKM
|
phosphofructokinase, muscle
|
|
chr14_+_51240205
Show fit
|
0.29 |
ENST00000457354.7
|
TMX1
|
thioredoxin related transmembrane protein 1
|
|
chr17_+_29568027
Show fit
|
0.28 |
ENST00000580183.6
ENST00000578749.5
ENST00000582829.6
|
TP53I13
|
tumor protein p53 inducible protein 13
|
|
chr21_+_38272410
Show fit
|
0.28 |
ENST00000398934.5
ENST00000398930.5
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15
|
|
chr20_+_43667105
Show fit
|
0.28 |
ENST00000217026.5
|
MYBL2
|
MYB proto-oncogene like 2
|
|
chrX_+_69504320
Show fit
|
0.27 |
ENST00000252338.5
|
FAM155B
|
family with sequence similarity 155 member B
|
|
chr18_-_51197671
Show fit
|
0.27 |
ENST00000406189.4
|
MEX3C
|
mex-3 RNA binding family member C
|
|
chr22_+_37675629
Show fit
|
0.27 |
ENST00000215909.10
|
LGALS1
|
galectin 1
|
|
chr13_-_113208087
Show fit
|
0.27 |
ENST00000375459.5
|
PCID2
|
PCI domain containing 2
|
|
chr11_+_126355634
Show fit
|
0.27 |
ENST00000227495.10
ENST00000676545.1
ENST00000678865.1
ENST00000444328.7
ENST00000677503.1
|
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4
|
|
chr1_-_32362081
Show fit
|
0.27 |
ENST00000432622.1
|
FAM229A
|
family with sequence similarity 229 member A
|
|
chrX_-_130165825
Show fit
|
0.27 |
ENST00000675240.1
ENST00000319908.8
ENST00000674546.1
ENST00000287295.8
|
AIFM1
|
apoptosis inducing factor mitochondria associated 1
|
|
chr12_-_6688859
Show fit
|
0.27 |
ENST00000542351.5
ENST00000538829.5
|
ZNF384
|
zinc finger protein 384
|
|
chr1_-_111140040
Show fit
|
0.27 |
ENST00000286692.8
ENST00000484310.6
|
DRAM2
|
DNA damage regulated autophagy modulator 2
|
|
chrX_-_123733023
Show fit
|
0.27 |
ENST00000245838.13
ENST00000355725.8
|
THOC2
|
THO complex 2
|
|
chr14_-_23365149
Show fit
|
0.26 |
ENST00000216733.8
|
EFS
|
embryonal Fyn-associated substrate
|
|
chr8_-_56074492
Show fit
|
0.26 |
ENST00000518875.5
ENST00000524349.5
ENST00000009589.8
ENST00000519606.5
ENST00000519807.5
ENST00000520627.1
ENST00000521262.5
|
RPS20
|
ribosomal protein S20
|
|
chr6_+_24356903
Show fit
|
0.26 |
ENST00000274766.2
|
KAAG1
|
kidney associated antigen 1
|
|
chr8_+_27774530
Show fit
|
0.26 |
ENST00000305188.13
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2
|
|
chr4_-_99088704
Show fit
|
0.26 |
ENST00000626055.2
ENST00000296412.14
|
ADH5
|
alcohol dehydrogenase 5 (class III), chi polypeptide
|
|
chr7_+_2244476
Show fit
|
0.25 |
ENST00000397049.2
|
NUDT1
|
nudix hydrolase 1
|
|
chr10_-_119872754
Show fit
|
0.25 |
ENST00000360003.7
|
MCMBP
|
minichromosome maintenance complex binding protein
|
|
chr1_+_16921923
Show fit
|
0.25 |
ENST00000375541.10
|
CROCC
|
ciliary rootlet coiled-coil, rootletin
|
|
chr7_-_141014939
Show fit
|
0.25 |
ENST00000324787.10
ENST00000467334.1
|
MRPS33
|
mitochondrial ribosomal protein S33
|
|
chr7_+_45157784
Show fit
|
0.25 |
ENST00000242249.8
ENST00000496212.5
ENST00000481345.1
|
RAMP3
|
receptor activity modifying protein 3
|
|
chr4_+_184649716
Show fit
|
0.25 |
ENST00000512834.5
ENST00000314970.11
|
PRIMPOL
|
primase and DNA directed polymerase
|
|
chr11_+_126355894
Show fit
|
0.24 |
ENST00000530591.5
ENST00000534083.5
|
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4
|
|
chr12_-_16608183
Show fit
|
0.24 |
ENST00000354662.5
ENST00000538051.5
|
LMO3
|
LIM domain only 3
|
|
chr2_-_37324826
Show fit
|
0.24 |
ENST00000234179.8
|
PRKD3
|
protein kinase D3
|
|
chr11_+_13009306
Show fit
|
0.24 |
ENST00000529419.3
|
RASSF10
|
Ras association domain family member 10
|
|
chr2_+_24123454
Show fit
|
0.24 |
ENST00000615575.5
|
FAM228B
|
family with sequence similarity 228 member B
|
|
chr18_-_32470872
Show fit
|
0.24 |
ENST00000269209.7
|
GAREM1
|
GRB2 associated regulator of MAPK1 subtype 1
|
|
chr1_-_6235945
Show fit
|
0.24 |
ENST00000343813.10
|
ICMT
|
isoprenylcysteine carboxyl methyltransferase
|
|
chrX_+_134237047
Show fit
|
0.23 |
ENST00000370809.4
ENST00000517294.5
|
CCDC160
|
coiled-coil domain containing 160
|
|
chr1_+_88684513
Show fit
|
0.23 |
ENST00000370513.9
|
PKN2
|
protein kinase N2
|
|
chr2_-_86563349
Show fit
|
0.23 |
ENST00000409727.5
|
CHMP3
|
charged multivesicular body protein 3
|
|
chr12_+_123671105
Show fit
|
0.23 |
ENST00000680574.1
ENST00000426174.6
ENST00000679504.1
ENST00000303372.7
|
TCTN2
|
tectonic family member 2
|
|
chr1_-_23980278
Show fit
|
0.23 |
ENST00000374453.7
ENST00000453840.7
|
SRSF10
|
serine and arginine rich splicing factor 10
|
|
chr6_-_26659685
Show fit
|
0.23 |
ENST00000480036.5
ENST00000415922.7
ENST00000622479.4
ENST00000607204.5
ENST00000456172.5
|
ZNF322
|
zinc finger protein 322
|
|
chr3_+_197950176
Show fit
|
0.23 |
ENST00000448864.6
ENST00000647248.2
|
RPL35A
|
ribosomal protein L35a
|
|
chr1_-_43172244
Show fit
|
0.23 |
ENST00000236051.3
|
EBNA1BP2
|
EBNA1 binding protein 2
|
|
chr12_-_48105808
Show fit
|
0.22 |
ENST00000448372.5
|
SENP1
|
SUMO specific peptidase 1
|
|
chr3_-_79767987
Show fit
|
0.22 |
ENST00000464233.6
|
ROBO1
|
roundabout guidance receptor 1
|
|
chr4_-_128288163
Show fit
|
0.22 |
ENST00000512483.5
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chr10_-_119872832
Show fit
|
0.22 |
ENST00000369077.4
|
MCMBP
|
minichromosome maintenance complex binding protein
|
|
chr12_-_54588636
Show fit
|
0.22 |
ENST00000257905.13
|
PPP1R1A
|
protein phosphatase 1 regulatory inhibitor subunit 1A
|
|
chr8_-_91040814
Show fit
|
0.22 |
ENST00000520014.1
ENST00000285419.8
|
PIP4P2
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 2
|
|
chr6_-_165655303
Show fit
|
0.22 |
ENST00000648917.1
|
PDE10A
|
phosphodiesterase 10A
|