Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
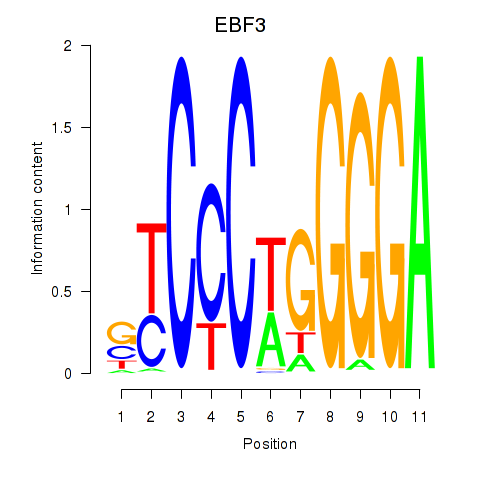
Results for EBF3
Z-value: 1.40
Transcription factors associated with EBF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EBF3
|
ENSG00000108001.16 | EBF transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EBF3 | hg38_v1_chr10_-_129964240_129964291 | 0.40 | 2.5e-02 | Click! |
Activity profile of EBF3 motif
Sorted Z-values of EBF3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_106335613 | 6.68 |
ENST00000603660.1
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr2_-_89143133 | 4.18 |
ENST00000492167.1
|
IGKV3-20
|
immunoglobulin kappa variable 3-20 |
| chr14_-_106269133 | 4.07 |
ENST00000390609.3
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr14_-_106360320 | 4.05 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
| chr22_+_22880706 | 3.99 |
ENST00000390319.2
|
IGLV3-1
|
immunoglobulin lambda variable 3-1 |
| chr19_+_41877267 | 3.81 |
ENST00000221972.8
ENST00000597454.1 ENST00000444740.2 |
CD79A
|
CD79a molecule |
| chr14_-_106235582 | 3.80 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr2_-_89177160 | 3.57 |
ENST00000484817.1
|
IGKV2-24
|
immunoglobulin kappa variable 2-24 |
| chr14_-_106062670 | 3.46 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr14_-_105588322 | 3.38 |
ENST00000497872.4
ENST00000390539.2 |
IGHA2
|
immunoglobulin heavy constant alpha 2 (A2m marker) |
| chr22_+_22887780 | 3.34 |
ENST00000532223.2
ENST00000526893.6 ENST00000531372.1 |
IGLL5
|
immunoglobulin lambda like polypeptide 5 |
| chr14_-_106538331 | 3.20 |
ENST00000390624.3
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr5_-_139389905 | 3.07 |
ENST00000302125.9
|
MZB1
|
marginal zone B and B1 cell specific protein |
| chr4_+_42397473 | 2.99 |
ENST00000319234.5
|
SHISA3
|
shisa family member 3 |
| chr2_-_89027700 | 2.92 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11 |
| chr14_-_106130061 | 2.82 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr2_+_89947508 | 2.65 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr2_+_90004792 | 2.63 |
ENST00000462693.1
|
IGKV2D-24
|
immunoglobulin kappa variable 2D-24 (non-functional) |
| chr14_-_106803221 | 2.56 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr14_-_106117159 | 2.51 |
ENST00000390601.3
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 |
| chr15_-_40340900 | 2.49 |
ENST00000559313.5
|
CCDC9B
|
coiled-coil domain containing 9B |
| chr11_+_60455839 | 2.48 |
ENST00000532491.5
ENST00000532073.5 ENST00000345732.9 ENST00000534668.6 ENST00000528313.1 ENST00000533306.6 ENST00000674194.1 |
MS4A1
|
membrane spanning 4-domains A1 |
| chr3_-_46882106 | 2.47 |
ENST00000662933.1
|
MYL3
|
myosin light chain 3 |
| chr3_-_142889156 | 2.45 |
ENST00000648195.1
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr3_-_142889075 | 2.42 |
ENST00000295992.8
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr16_+_32066065 | 2.38 |
ENST00000354689.6
|
IGHV3OR16-9
|
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr7_-_44141285 | 2.34 |
ENST00000458240.5
ENST00000223364.7 |
MYL7
|
myosin light chain 7 |
| chr1_-_153544997 | 2.34 |
ENST00000368715.5
|
S100A4
|
S100 calcium binding protein A4 |
| chr7_-_44141074 | 2.15 |
ENST00000457314.5
ENST00000447951.1 ENST00000431007.1 |
MYL7
|
myosin light chain 7 |
| chr6_-_46015812 | 2.10 |
ENST00000544153.3
ENST00000339561.12 |
CLIC5
|
chloride intracellular channel 5 |
| chr3_-_142888896 | 2.10 |
ENST00000485766.1
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr19_-_45322867 | 2.10 |
ENST00000221476.4
|
CKM
|
creatine kinase, M-type |
| chr19_-_38617928 | 2.05 |
ENST00000396857.7
ENST00000586296.5 |
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr7_-_150632333 | 2.02 |
ENST00000493969.2
ENST00000328902.9 |
GIMAP6
|
GTPase, IMAP family member 6 |
| chr7_-_150632644 | 1.98 |
ENST00000618759.4
|
GIMAP6
|
GTPase, IMAP family member 6 |
| chr7_+_80624071 | 1.98 |
ENST00000438020.5
|
CD36
|
CD36 molecule |
| chr11_-_102874974 | 1.97 |
ENST00000571244.3
|
MMP12
|
matrix metallopeptidase 12 |
| chr20_+_31637905 | 1.97 |
ENST00000376075.4
|
COX4I2
|
cytochrome c oxidase subunit 4I2 |
| chr2_+_87338511 | 1.95 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr19_-_7702139 | 1.88 |
ENST00000346664.9
|
FCER2
|
Fc fragment of IgE receptor II |
| chr10_+_86668501 | 1.86 |
ENST00000623056.4
ENST00000263066.11 ENST00000429277.7 ENST00000361373.9 ENST00000372066.8 ENST00000372056.8 ENST00000623007.3 |
LDB3
|
LIM domain binding 3 |
| chr19_-_7702124 | 1.85 |
ENST00000597921.6
|
FCER2
|
Fc fragment of IgE receptor II |
| chr14_-_106165730 | 1.81 |
ENST00000390604.2
|
IGHV3-16
|
immunoglobulin heavy variable 3-16 (non-functional) |
| chr14_-_106088573 | 1.81 |
ENST00000632099.1
|
IGHV3-64D
|
immunoglobulin heavy variable 3-64D |
| chr19_+_17527250 | 1.80 |
ENST00000599164.6
ENST00000449408.6 ENST00000600871.5 ENST00000599124.1 |
NIBAN3
|
niban apoptosis regulator 3 |
| chr2_-_187513641 | 1.80 |
ENST00000392365.5
ENST00000435414.5 |
TFPI
|
tissue factor pathway inhibitor |
| chr12_+_119178920 | 1.80 |
ENST00000281938.7
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr19_-_38617912 | 1.71 |
ENST00000591517.5
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr14_-_106791226 | 1.70 |
ENST00000433072.2
|
IGHV3-72
|
immunoglobulin heavy variable 3-72 |
| chr15_+_73926443 | 1.63 |
ENST00000261921.8
|
LOXL1
|
lysyl oxidase like 1 |
| chr14_-_89494051 | 1.63 |
ENST00000555034.5
ENST00000553904.1 |
FOXN3
|
forkhead box N3 |
| chr22_-_37244237 | 1.59 |
ENST00000401529.3
ENST00000249071.11 |
RAC2
|
Rac family small GTPase 2 |
| chr12_+_56994485 | 1.57 |
ENST00000300098.3
|
GPR182
|
G protein-coupled receptor 182 |
| chr6_-_46015607 | 1.54 |
ENST00000644878.1
ENST00000644324.1 ENST00000672327.1 |
CLIC5
|
chloride intracellular channel 5 |
| chr1_-_153550083 | 1.49 |
ENST00000368714.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr8_+_141417902 | 1.46 |
ENST00000681443.1
|
PTP4A3
|
protein tyrosine phosphatase 4A3 |
| chrX_+_129779930 | 1.46 |
ENST00000356892.4
|
SASH3
|
SAM and SH3 domain containing 3 |
| chr2_+_158968608 | 1.44 |
ENST00000263635.8
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr12_+_119178953 | 1.40 |
ENST00000674542.1
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr11_-_111910888 | 1.40 |
ENST00000525823.1
ENST00000528961.6 |
CRYAB
|
crystallin alpha B |
| chr19_+_3933059 | 1.37 |
ENST00000616156.4
ENST00000168977.6 ENST00000599576.5 |
NMRK2
|
nicotinamide riboside kinase 2 |
| chr3_-_119660580 | 1.36 |
ENST00000493094.6
ENST00000264231.7 ENST00000468801.1 |
POPDC2
|
popeye domain containing 2 |
| chr3_-_46882165 | 1.35 |
ENST00000431168.1
ENST00000654597.1 |
MYL3
|
myosin light chain 3 |
| chr22_-_46537593 | 1.35 |
ENST00000262738.9
ENST00000674500.2 |
CELSR1
|
cadherin EGF LAG seven-pass G-type receptor 1 |
| chr17_+_1279655 | 1.32 |
ENST00000333813.4
|
TRARG1
|
trafficking regulator of GLUT4 (SLC2A4) 1 |
| chr8_-_19602484 | 1.31 |
ENST00000454498.6
ENST00000520003.5 |
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr19_-_15464525 | 1.30 |
ENST00000343625.12
|
RASAL3
|
RAS protein activator like 3 |
| chr5_+_149271848 | 1.29 |
ENST00000296721.9
|
AFAP1L1
|
actin filament associated protein 1 like 1 |
| chr19_+_17527232 | 1.28 |
ENST00000601861.5
|
NIBAN3
|
niban apoptosis regulator 3 |
| chr17_-_1492660 | 1.27 |
ENST00000648651.1
|
MYO1C
|
myosin IC |
| chr2_+_102761963 | 1.26 |
ENST00000640575.2
ENST00000412401.3 |
TMEM182
|
transmembrane protein 182 |
| chr5_-_41510554 | 1.26 |
ENST00000377801.8
|
PLCXD3
|
phosphatidylinositol specific phospholipase C X domain containing 3 |
| chr11_+_2444986 | 1.24 |
ENST00000155840.12
|
KCNQ1
|
potassium voltage-gated channel subfamily Q member 1 |
| chr6_-_32224060 | 1.23 |
ENST00000375023.3
|
NOTCH4
|
notch receptor 4 |
| chr1_-_24112125 | 1.23 |
ENST00000374434.4
|
MYOM3
|
myomesin 3 |
| chr20_-_64049631 | 1.23 |
ENST00000340356.9
|
SOX18
|
SRY-box transcription factor 18 |
| chrX_+_136147525 | 1.23 |
ENST00000652745.1
ENST00000627578.2 ENST00000652457.1 ENST00000394155.8 ENST00000618438.4 |
FHL1
|
four and a half LIM domains 1 |
| chr17_+_79024142 | 1.22 |
ENST00000579760.6
ENST00000339142.6 |
C1QTNF1
|
C1q and TNF related 1 |
| chr5_+_149271906 | 1.22 |
ENST00000515000.1
|
AFAP1L1
|
actin filament associated protein 1 like 1 |
| chr2_+_89985922 | 1.21 |
ENST00000390268.2
|
IGKV2D-26
|
immunoglobulin kappa variable 2D-26 |
| chr14_+_105486867 | 1.18 |
ENST00000409393.6
ENST00000392531.4 |
CRIP1
|
cysteine rich protein 1 |
| chr16_-_33845229 | 1.16 |
ENST00000569103.2
|
IGHV3OR16-17
|
immunoglobulin heavy variable 3/OR16-17 (non-functional) |
| chrX_+_136147465 | 1.16 |
ENST00000651929.2
|
FHL1
|
four and a half LIM domains 1 |
| chr19_+_39413528 | 1.15 |
ENST00000438123.5
ENST00000409797.6 ENST00000451354.6 |
PLEKHG2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr5_+_120464236 | 1.15 |
ENST00000407149.7
ENST00000379551.2 |
PRR16
|
proline rich 16 |
| chr14_-_106470788 | 1.13 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr17_+_79025612 | 1.12 |
ENST00000392445.6
|
C1QTNF1
|
C1q and TNF related 1 |
| chrX_+_136147556 | 1.12 |
ENST00000651089.1
ENST00000420362.5 |
FHL1
|
four and a half LIM domains 1 |
| chr22_-_37244417 | 1.11 |
ENST00000405484.5
ENST00000441619.5 ENST00000406508.5 |
RAC2
|
Rac family small GTPase 2 |
| chrX_+_136205982 | 1.10 |
ENST00000628568.1
|
FHL1
|
four and a half LIM domains 1 |
| chr11_+_75562242 | 1.10 |
ENST00000526397.5
ENST00000529643.1 ENST00000525492.5 ENST00000530284.5 ENST00000358171.8 |
SERPINH1
|
serpin family H member 1 |
| chr9_-_114348966 | 1.10 |
ENST00000374079.8
|
AKNA
|
AT-hook transcription factor |
| chr10_+_92689946 | 1.09 |
ENST00000282728.10
|
HHEX
|
hematopoietically expressed homeobox |
| chr15_+_63042632 | 1.08 |
ENST00000288398.10
ENST00000358278.7 ENST00000610733.1 ENST00000403994.9 ENST00000357980.9 ENST00000559556.5 ENST00000267996.11 ENST00000559397.6 ENST00000561266.6 ENST00000560970.6 |
TPM1
|
tropomyosin 1 |
| chr16_+_32995048 | 1.08 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr14_+_105486311 | 1.08 |
ENST00000330233.11
|
CRIP1
|
cysteine rich protein 1 |
| chr5_-_41510623 | 1.08 |
ENST00000328457.5
|
PLCXD3
|
phosphatidylinositol specific phospholipase C X domain containing 3 |
| chr6_-_41940537 | 1.07 |
ENST00000512426.5
|
CCND3
|
cyclin D3 |
| chr20_-_50636887 | 1.07 |
ENST00000045083.6
|
RIPOR3
|
RIPOR family member 3 |
| chr3_-_16512895 | 1.07 |
ENST00000449415.5
ENST00000441460.5 |
RFTN1
|
raftlin, lipid raft linker 1 |
| chr22_+_36860973 | 1.06 |
ENST00000447071.5
ENST00000397147.7 ENST00000248899.11 |
NCF4
|
neutrophil cytosolic factor 4 |
| chr19_+_3933581 | 1.06 |
ENST00000593949.1
|
NMRK2
|
nicotinamide riboside kinase 2 |
| chr6_-_7910776 | 1.06 |
ENST00000379757.9
|
TXNDC5
|
thioredoxin domain containing 5 |
| chr15_+_90184912 | 1.05 |
ENST00000561085.1
ENST00000332496.10 |
SEMA4B
|
semaphorin 4B |
| chr14_-_53951880 | 1.04 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr11_+_64234569 | 1.04 |
ENST00000309422.7
ENST00000426086.3 |
VEGFB
|
vascular endothelial growth factor B |
| chr10_+_97572771 | 1.03 |
ENST00000370655.6
ENST00000455090.1 |
ANKRD2
|
ankyrin repeat domain 2 |
| chr10_-_123891742 | 1.01 |
ENST00000241305.4
ENST00000615851.4 |
CPXM2
|
carboxypeptidase X, M14 family member 2 |
| chr15_-_19988117 | 1.01 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr17_+_42854078 | 1.00 |
ENST00000591562.1
ENST00000588033.1 |
AOC3
|
amine oxidase copper containing 3 |
| chr11_+_75562274 | 1.00 |
ENST00000532356.5
ENST00000524558.5 |
SERPINH1
|
serpin family H member 1 |
| chr17_+_79024243 | 0.99 |
ENST00000311661.4
|
C1QTNF1
|
C1q and TNF related 1 |
| chr17_+_75610690 | 0.99 |
ENST00000642007.1
|
MYO15B
|
myosin XVB |
| chr3_-_49132994 | 0.98 |
ENST00000305544.9
ENST00000494831.1 ENST00000418109.5 |
LAMB2
|
laminin subunit beta 2 |
| chr18_+_31498168 | 0.98 |
ENST00000261590.13
ENST00000585206.1 ENST00000683654.1 |
DSG2
|
desmoglein 2 |
| chr5_-_176537361 | 0.97 |
ENST00000274811.9
|
RNF44
|
ring finger protein 44 |
| chr2_-_110116022 | 0.97 |
ENST00000427178.1
|
MALL
|
mal, T cell differentiation protein like |
| chrX_+_118495803 | 0.96 |
ENST00000276202.9
ENST00000276204.10 |
DOCK11
|
dedicator of cytokinesis 11 |
| chr1_+_159171607 | 0.95 |
ENST00000368124.8
ENST00000368125.9 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr10_-_73655984 | 0.95 |
ENST00000394810.3
|
SYNPO2L
|
synaptopodin 2 like |
| chr7_+_76303547 | 0.95 |
ENST00000429938.1
|
HSPB1
|
heat shock protein family B (small) member 1 |
| chr10_+_30434021 | 0.94 |
ENST00000542547.5
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr2_+_48530132 | 0.94 |
ENST00000404752.6
ENST00000406226.1 |
STON1
|
stonin 1 |
| chr19_-_2042066 | 0.94 |
ENST00000591588.1
ENST00000591142.5 |
MKNK2
|
MAPK interacting serine/threonine kinase 2 |
| chr11_+_75562056 | 0.93 |
ENST00000533603.5
|
SERPINH1
|
serpin family H member 1 |
| chr1_-_178869272 | 0.93 |
ENST00000444255.1
|
ANGPTL1
|
angiopoietin like 1 |
| chr11_+_62270150 | 0.92 |
ENST00000525380.1
ENST00000227918.3 |
SCGB2A2
|
secretoglobin family 2A member 2 |
| chr5_-_172771187 | 0.91 |
ENST00000239223.4
|
DUSP1
|
dual specificity phosphatase 1 |
| chr2_+_46542474 | 0.90 |
ENST00000238738.9
|
RHOQ
|
ras homolog family member Q |
| chr10_+_97572493 | 0.89 |
ENST00000307518.9
ENST00000298808.9 |
ANKRD2
|
ankyrin repeat domain 2 |
| chr19_+_45507470 | 0.88 |
ENST00000245932.11
ENST00000592139.1 ENST00000590603.1 |
VASP
|
vasodilator stimulated phosphoprotein |
| chr1_-_154970735 | 0.86 |
ENST00000368445.9
ENST00000448116.7 ENST00000368449.8 |
SHC1
|
SHC adaptor protein 1 |
| chr6_-_106974721 | 0.86 |
ENST00000606017.2
ENST00000620405.1 |
CD24
|
CD24 molecule |
| chr5_-_141878396 | 0.85 |
ENST00000503492.5
ENST00000287008.8 ENST00000394536.4 |
PCDH1
|
protocadherin 1 |
| chr5_-_38556625 | 0.85 |
ENST00000506990.5
ENST00000453190.7 |
LIFR
|
LIF receptor subunit alpha |
| chr3_-_88059042 | 0.83 |
ENST00000309534.10
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr19_-_6591103 | 0.83 |
ENST00000423145.7
ENST00000245903.4 |
CD70
|
CD70 molecule |
| chr10_-_95561355 | 0.82 |
ENST00000607232.5
ENST00000371247.7 ENST00000371227.8 ENST00000371249.6 ENST00000371246.6 ENST00000306402.10 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr1_+_26543106 | 0.82 |
ENST00000530003.5
|
RPS6KA1
|
ribosomal protein S6 kinase A1 |
| chr8_+_141128612 | 0.81 |
ENST00000518347.5
ENST00000262585.6 ENST00000520986.5 ENST00000523058.5 ENST00000518668.5 |
DENND3
|
DENN domain containing 3 |
| chr11_+_20022829 | 0.81 |
ENST00000525322.5
ENST00000530408.1 |
NAV2
|
neuron navigator 2 |
| chr1_+_2556361 | 0.76 |
ENST00000355716.5
|
TNFRSF14
|
TNF receptor superfamily member 14 |
| chr5_+_140175117 | 0.76 |
ENST00000261811.6
|
CYSTM1
|
cysteine rich transmembrane module containing 1 |
| chrX_+_154420315 | 0.76 |
ENST00000651139.1
|
TAZ
|
tafazzin |
| chr21_-_38661694 | 0.76 |
ENST00000417133.6
ENST00000398910.5 ENST00000442448.5 ENST00000429727.6 |
ERG
|
ETS transcription factor ERG |
| chr11_+_20022550 | 0.76 |
ENST00000533917.5
|
NAV2
|
neuron navigator 2 |
| chr10_-_13972355 | 0.75 |
ENST00000264546.10
|
FRMD4A
|
FERM domain containing 4A |
| chr8_+_141128581 | 0.74 |
ENST00000519811.6
|
DENND3
|
DENN domain containing 3 |
| chr19_-_40266315 | 0.72 |
ENST00000441941.6
ENST00000580747.5 |
AKT2
|
AKT serine/threonine kinase 2 |
| chr17_-_80220325 | 0.71 |
ENST00000326317.11
ENST00000570427.1 ENST00000570923.1 |
SGSH
|
N-sulfoglucosamine sulfohydrolase |
| chr10_+_30434176 | 0.71 |
ENST00000263056.6
ENST00000375322.2 |
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr2_+_201233443 | 0.71 |
ENST00000392263.6
ENST00000264274.13 ENST00000432109.6 ENST00000264275.9 ENST00000450491.5 ENST00000440732.5 ENST00000392258.7 |
CASP8
|
caspase 8 |
| chr18_+_11981428 | 0.71 |
ENST00000625802.2
|
IMPA2
|
inositol monophosphatase 2 |
| chr19_+_40601342 | 0.70 |
ENST00000396819.8
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr3_-_88058928 | 0.69 |
ENST00000482016.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr17_+_2056073 | 0.69 |
ENST00000576444.1
ENST00000322941.3 |
HIC1
|
HIC ZBTB transcriptional repressor 1 |
| chr10_+_30434116 | 0.69 |
ENST00000415139.5
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr10_-_28303051 | 0.69 |
ENST00000683449.1
|
MPP7
|
membrane palmitoylated protein 7 |
| chr4_+_986997 | 0.69 |
ENST00000247933.9
|
IDUA
|
alpha-L-iduronidase |
| chr3_+_141368497 | 0.69 |
ENST00000321464.7
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr18_+_34493289 | 0.69 |
ENST00000682923.1
ENST00000596745.5 ENST00000283365.14 ENST00000315456.10 ENST00000598774.6 ENST00000684266.1 ENST00000683092.1 ENST00000683379.1 ENST00000684359.1 |
DTNA
|
dystrobrevin alpha |
| chr4_+_987037 | 0.68 |
ENST00000502910.5
ENST00000514224.2 ENST00000504568.5 |
IDUA
|
alpha-L-iduronidase |
| chr6_-_41940690 | 0.68 |
ENST00000505064.1
|
CCND3
|
cyclin D3 |
| chr9_-_35619542 | 0.67 |
ENST00000396757.6
|
CD72
|
CD72 molecule |
| chr12_-_47758645 | 0.67 |
ENST00000395358.7
|
RAPGEF3
|
Rap guanine nucleotide exchange factor 3 |
| chr10_+_17809337 | 0.67 |
ENST00000569591.3
|
MRC1
|
mannose receptor C-type 1 |
| chr6_-_29681112 | 0.66 |
ENST00000376883.2
ENST00000488757.6 |
ZFP57
|
ZFP57 zinc finger protein |
| chr9_+_132582631 | 0.66 |
ENST00000542090.1
|
BARHL1
|
BarH like homeobox 1 |
| chr12_-_47758828 | 0.66 |
ENST00000389212.7
ENST00000449771.7 |
RAPGEF3
|
Rap guanine nucleotide exchange factor 3 |
| chr2_+_27078598 | 0.65 |
ENST00000380320.9
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr11_+_124865425 | 0.65 |
ENST00000397801.6
|
ROBO3
|
roundabout guidance receptor 3 |
| chr5_+_136047319 | 0.62 |
ENST00000508767.5
ENST00000604555.5 |
TGFBI
|
transforming growth factor beta induced |
| chr1_+_2556222 | 0.61 |
ENST00000409119.5
|
TNFRSF14
|
TNF receptor superfamily member 14 |
| chr21_-_14658812 | 0.61 |
ENST00000647101.1
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr16_+_81739027 | 0.61 |
ENST00000566191.5
ENST00000565272.5 ENST00000563954.5 ENST00000565054.5 |
ENSG00000261218.5
PLCG2
|
novel transcript phospholipase C gamma 2 |
| chr8_+_12945667 | 0.61 |
ENST00000524591.7
|
TRMT9B
|
tRNA methyltransferase 9B (putative) |
| chr7_-_44979003 | 0.61 |
ENST00000258787.12
ENST00000648014.1 |
MYO1G
|
myosin IG |
| chr8_-_25424260 | 0.60 |
ENST00000421054.7
|
GNRH1
|
gonadotropin releasing hormone 1 |
| chr14_+_54567612 | 0.60 |
ENST00000251091.9
ENST00000392067.7 ENST00000631086.2 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr5_+_15500172 | 0.60 |
ENST00000504595.2
|
FBXL7
|
F-box and leucine rich repeat protein 7 |
| chr11_-_33722941 | 0.60 |
ENST00000652678.1
|
CD59
|
CD59 molecule (CD59 blood group) |
| chr10_+_110644306 | 0.60 |
ENST00000369519.4
|
RBM20
|
RNA binding motif protein 20 |
| chr19_+_51689128 | 0.59 |
ENST00000646845.1
|
SPACA6
|
sperm acrosome associated 6 |
| chrX_-_155026868 | 0.59 |
ENST00000453950.1
ENST00000423959.5 |
F8
|
coagulation factor VIII |
| chr5_+_134525649 | 0.59 |
ENST00000282605.8
ENST00000681547.2 ENST00000361895.6 ENST00000402835.5 |
JADE2
|
jade family PHD finger 2 |
| chr7_-_27147774 | 0.59 |
ENST00000222728.3
|
HOXA6
|
homeobox A6 |
| chr3_-_10292933 | 0.59 |
ENST00000429122.1
ENST00000425479.1 ENST00000335542.13 |
GHRL
|
ghrelin and obestatin prepropeptide |
| chr9_+_132582581 | 0.59 |
ENST00000263610.7
|
BARHL1
|
BarH like homeobox 1 |
| chr4_-_36243939 | 0.59 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr17_+_49210387 | 0.57 |
ENST00000419580.6
|
ABI3
|
ABI family member 3 |
| chr4_+_4859658 | 0.57 |
ENST00000382723.5
|
MSX1
|
msh homeobox 1 |
| chr4_-_128287785 | 0.57 |
ENST00000296425.10
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr20_+_2692736 | 0.57 |
ENST00000380648.9
ENST00000497450.5 |
EBF4
|
EBF family member 4 |
| chr4_-_128288163 | 0.56 |
ENST00000512483.5
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr20_-_57709760 | 0.56 |
ENST00000395819.3
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr16_-_11976611 | 0.56 |
ENST00000538896.5
ENST00000673243.1 |
NPIPB2
|
nuclear pore complex interacting protein family member B2 |
| chr15_+_75347610 | 0.56 |
ENST00000564784.5
ENST00000569035.5 |
NEIL1
|
nei like DNA glycosylase 1 |
| chr6_-_30687200 | 0.56 |
ENST00000399199.7
|
PPP1R18
|
protein phosphatase 1 regulatory subunit 18 |
| chr5_+_140175205 | 0.56 |
ENST00000644078.1
|
CYSTM1
|
cysteine rich transmembrane module containing 1 |
| chr7_-_82443766 | 0.56 |
ENST00000356860.8
|
CACNA2D1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EBF3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.5 | 2.0 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.5 | 3.3 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.5 | 3.7 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.4 | 1.6 | GO:0018277 | protein deamination(GO:0018277) |
| 0.4 | 1.1 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.4 | 1.1 | GO:2000506 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) negative regulation of energy homeostasis(GO:2000506) |
| 0.4 | 60.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.3 | 3.7 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.3 | 1.3 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.3 | 1.0 | GO:0072313 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.3 | 2.4 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.3 | 3.2 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.3 | 1.3 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.2 | 1.0 | GO:0003165 | Purkinje myocyte development(GO:0003165) |
| 0.2 | 2.6 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) |
| 0.2 | 1.3 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.2 | 1.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.2 | 0.9 | GO:0032597 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.2 | 1.0 | GO:0072101 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.2 | 1.2 | GO:0060214 | stem cell fate commitment(GO:0048865) endocardium formation(GO:0060214) |
| 0.2 | 0.6 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.2 | 2.0 | GO:0072642 | interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.2 | 0.6 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.2 | 1.7 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 0.7 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.2 | 1.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 0.5 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.2 | 0.3 | GO:0061027 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) |
| 0.2 | 2.2 | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway(GO:0038089) |
| 0.2 | 0.5 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.2 | 1.0 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.2 | 4.0 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.2 | 2.0 | GO:2000334 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 0.6 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 2.5 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.6 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.7 | GO:0039516 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.1 | 0.8 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 1.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.4 | GO:1904828 | regulation of phenotypic switching by transcription from RNA polymerase II promoter(GO:0100057) regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.1 | 1.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 4.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.4 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.1 | 0.8 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 1.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.6 | GO:2000330 | interleukin-23-mediated signaling pathway(GO:0038155) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.1 | 1.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.6 | GO:0001971 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.1 | 0.7 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.7 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.3 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.1 | 0.7 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.1 | 3.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.5 | GO:1904141 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.1 | 0.5 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 1.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 1.2 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 0.4 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 0.9 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.7 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.2 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.1 | 1.4 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 2.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 1.4 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.1 | 0.2 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.3 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.1 | 0.9 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 0.8 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.1 | 0.5 | GO:0010814 | neuropeptide catabolic process(GO:0010813) substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 1.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.3 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.7 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.3 | GO:1902904 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.1 | 4.9 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 0.2 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.4 | GO:0046985 | negative regulation of megakaryocyte differentiation(GO:0045653) positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 1.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 1.0 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.2 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.1 | 13.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.4 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.1 | 1.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.6 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.2 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.1 | 0.8 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 6.6 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 1.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.2 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.1 | 1.0 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.3 | GO:0070269 | ectopic germ cell programmed cell death(GO:0035234) pyroptosis(GO:0070269) |
| 0.0 | 0.2 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.8 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 4.6 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.7 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.2 | GO:1904306 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.0 | 0.8 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.4 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 1.4 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.2 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.5 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 1.1 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 1.0 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.5 | GO:0046641 | positive regulation of alpha-beta T cell proliferation(GO:0046641) |
| 0.0 | 0.6 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.3 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 6.0 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 1.8 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.6 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.2 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.6 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.8 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.4 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.4 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0000494 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.3 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.6 | GO:0060180 | female mating behavior(GO:0060180) |
| 0.0 | 1.0 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 1.8 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.6 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.4 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.4 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.8 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.9 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:1903778 | positive regulation of histamine secretion by mast cell(GO:1903595) protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.8 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 2.9 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.3 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 2.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 1.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.7 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.8 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.0 | 0.5 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.2 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.9 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.5 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.3 | GO:0031274 | regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.0 | 0.7 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.4 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.9 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.7 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.6 | GO:0072678 | T cell migration(GO:0072678) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0090294 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 2.9 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.2 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.9 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.2 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.0 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.3 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 1.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.9 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.0 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.6 | GO:0071745 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.8 | 3.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.4 | 22.8 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.3 | 1.0 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.3 | 1.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.2 | 0.6 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.2 | 3.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 0.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 0.8 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.1 | 1.3 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.1 | 2.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.9 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 5.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.7 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 1.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.5 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 5.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.3 | GO:0070701 | mucus layer(GO:0070701) |
| 0.1 | 0.4 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 0.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.3 | GO:0072557 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 1.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.3 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 2.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.6 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 2.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 2.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 2.5 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 9.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 1.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 1.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 2.0 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.7 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 3.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 2.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 3.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 4.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 3.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 3.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.5 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 4.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 2.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.4 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.8 | GO:0032432 | actin filament bundle(GO:0032432) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.6 | 3.8 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.6 | 2.4 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.4 | 3.7 | GO:0019863 | IgE binding(GO:0019863) |
| 0.4 | 26.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 1.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.3 | 0.9 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.3 | 1.3 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.3 | 2.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.7 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.2 | 0.9 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.2 | 0.9 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.2 | 0.6 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.2 | 0.8 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.2 | 1.0 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 0.6 | GO:0016517 | interleukin-12 receptor activity(GO:0016517) |
| 0.2 | 43.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.2 | 1.0 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.2 | 0.9 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 1.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.2 | 4.0 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 2.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 3.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 1.5 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 0.4 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.1 | 1.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 1.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.8 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.7 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 2.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.0 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.3 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.1 | 0.6 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 1.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.7 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 15.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.2 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 1.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.9 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 4.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.3 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.9 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.8 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.5 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 1.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 2.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 2.4 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 1.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.6 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.3 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 2.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 1.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.6 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 1.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 1.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.4 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.3 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 1.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 1.1 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.3 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 1.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.3 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.4 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 1.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.7 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.7 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 1.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 1.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 5.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 1.9 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 3.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 1.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.0 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.3 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 2.2 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 0.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 1.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 4.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 5.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 3.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.7 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 2.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.6 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 2.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 8.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 4.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.1 | 10.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 6.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.2 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 4.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 3.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 0.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 1.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 2.1 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 2.9 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 3.8 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 1.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.0 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |