Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
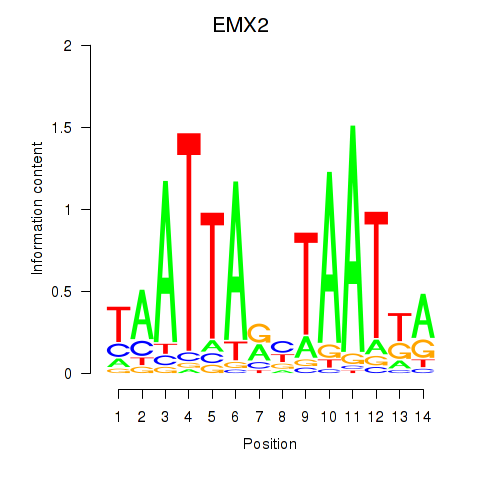
Results for EMX2
Z-value: 1.07
Transcription factors associated with EMX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EMX2
|
ENSG00000170370.12 | empty spiracles homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EMX2 | hg38_v1_chr10_+_117542416_117542445 | 0.15 | 4.2e-01 | Click! |
Activity profile of EMX2 motif
Sorted Z-values of EMX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_53962925 | 4.17 |
ENST00000270458.4
|
CACNG8
|
calcium voltage-gated channel auxiliary subunit gamma 8 |
| chr2_+_17541157 | 2.37 |
ENST00000406397.1
|
VSNL1
|
visinin like 1 |
| chr10_-_20897288 | 2.14 |
ENST00000377122.9
|
NEBL
|
nebulette |
| chr6_+_13272709 | 2.12 |
ENST00000379335.8
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr17_-_40937641 | 2.09 |
ENST00000209718.8
|
KRT23
|
keratin 23 |
| chr1_+_149782671 | 2.01 |
ENST00000444948.5
ENST00000369168.5 |
FCGR1A
|
Fc fragment of IgG receptor Ia |
| chr4_-_88284553 | 1.91 |
ENST00000608933.6
ENST00000295908.11 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chr4_-_88284616 | 1.89 |
ENST00000508256.5
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chr5_-_74866958 | 1.88 |
ENST00000389156.9
|
FAM169A
|
family with sequence similarity 169 member A |
| chr18_-_26865689 | 1.87 |
ENST00000675739.1
ENST00000383168.9 ENST00000672981.2 ENST00000578776.1 |
AQP4
|
aquaporin 4 |
| chr1_+_84181630 | 1.83 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr9_+_12695702 | 1.79 |
ENST00000381136.2
|
TYRP1
|
tyrosinase related protein 1 |
| chr5_+_36608146 | 1.77 |
ENST00000381918.4
ENST00000513646.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr17_-_40937445 | 1.76 |
ENST00000436344.7
ENST00000485751.1 |
KRT23
|
keratin 23 |
| chr4_-_88284590 | 1.74 |
ENST00000510548.6
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chrX_+_154304923 | 1.70 |
ENST00000426989.5
ENST00000426203.5 ENST00000369912.2 |
TKTL1
|
transketolase like 1 |
| chr18_-_26865732 | 1.53 |
ENST00000672188.1
|
AQP4
|
aquaporin 4 |
| chr16_+_85027735 | 1.52 |
ENST00000258180.7
ENST00000538274.5 |
KIAA0513
|
KIAA0513 |
| chr4_-_88284747 | 1.46 |
ENST00000514204.1
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chr16_-_46748337 | 1.45 |
ENST00000394809.9
|
MYLK3
|
myosin light chain kinase 3 |
| chr3_-_132037800 | 1.45 |
ENST00000617767.4
|
CPNE4
|
copine 4 |
| chr1_+_192809031 | 1.45 |
ENST00000235382.7
|
RGS2
|
regulator of G protein signaling 2 |
| chr12_+_59689337 | 1.40 |
ENST00000261187.8
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr1_+_84144260 | 1.32 |
ENST00000370685.7
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr6_+_72216442 | 1.27 |
ENST00000425662.6
ENST00000453976.6 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chrX_+_18425597 | 1.27 |
ENST00000623535.2
ENST00000674046.1 |
CDKL5
|
cyclin dependent kinase like 5 |
| chr5_-_138139382 | 1.26 |
ENST00000265191.4
|
NME5
|
NME/NM23 family member 5 |
| chr20_+_9514562 | 1.26 |
ENST00000246070.3
|
LAMP5
|
lysosomal associated membrane protein family member 5 |
| chr2_-_44323302 | 1.23 |
ENST00000420756.1
ENST00000444696.5 |
PREPL
|
prolyl endopeptidase like |
| chr3_-_196515315 | 1.23 |
ENST00000397537.3
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chrX_-_73214793 | 1.21 |
ENST00000373517.4
|
NAP1L2
|
nucleosome assembly protein 1 like 2 |
| chr17_-_59081162 | 1.18 |
ENST00000581468.1
|
TRIM37
|
tripartite motif containing 37 |
| chr3_-_151329539 | 1.16 |
ENST00000325602.6
|
P2RY13
|
purinergic receptor P2Y13 |
| chr5_-_11588842 | 1.15 |
ENST00000503622.5
|
CTNND2
|
catenin delta 2 |
| chrX_-_23907887 | 1.08 |
ENST00000379226.9
|
APOO
|
apolipoprotein O |
| chr2_-_2324323 | 1.06 |
ENST00000648339.1
ENST00000647694.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr12_-_86256299 | 1.06 |
ENST00000552808.6
ENST00000547225.5 |
MGAT4C
|
MGAT4 family member C |
| chr2_-_206159410 | 1.03 |
ENST00000457011.5
ENST00000440274.5 ENST00000432169.5 ENST00000233190.11 |
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr5_-_111976925 | 1.02 |
ENST00000395634.7
|
NREP
|
neuronal regeneration related protein |
| chr12_-_10130143 | 1.02 |
ENST00000298523.9
ENST00000396484.6 ENST00000310002.4 ENST00000304084.13 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr11_+_119149029 | 0.99 |
ENST00000619701.5
|
ABCG4
|
ATP binding cassette subfamily G member 4 |
| chr16_+_56191476 | 0.98 |
ENST00000262493.12
|
GNAO1
|
G protein subunit alpha o1 |
| chr10_+_27532521 | 0.98 |
ENST00000683924.1
|
RAB18
|
RAB18, member RAS oncogene family |
| chrX_+_18425579 | 0.97 |
ENST00000379996.7
|
CDKL5
|
cyclin dependent kinase like 5 |
| chr1_+_152878312 | 0.96 |
ENST00000368765.4
|
SMCP
|
sperm mitochondria associated cysteine rich protein |
| chr15_+_77420957 | 0.96 |
ENST00000559099.5
|
HMG20A
|
high mobility group 20A |
| chr2_-_208186999 | 0.94 |
ENST00000453017.5
ENST00000423952.2 |
C2orf80
|
chromosome 2 open reading frame 80 |
| chr5_-_74866914 | 0.93 |
ENST00000513277.5
|
FAM169A
|
family with sequence similarity 169 member A |
| chr8_-_73582830 | 0.93 |
ENST00000523533.5
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr14_+_24070837 | 0.92 |
ENST00000537691.5
ENST00000397016.6 ENST00000560356.5 ENST00000558450.5 |
CPNE6
|
copine 6 |
| chr2_+_227871618 | 0.91 |
ENST00000309931.3
ENST00000440997.1 |
DAW1
|
dynein assembly factor with WD repeats 1 |
| chr1_+_92168915 | 0.91 |
ENST00000637221.2
|
BTBD8
|
BTB domain containing 8 |
| chr6_-_110179855 | 0.90 |
ENST00000368938.5
|
WASF1
|
WASP family member 1 |
| chr6_-_110179995 | 0.89 |
ENST00000392586.5
ENST00000419252.1 ENST00000359451.6 ENST00000392588.5 |
WASF1
|
WASP family member 1 |
| chr1_+_178513103 | 0.89 |
ENST00000319416.7
ENST00000367643.7 ENST00000367642.3 ENST00000367641.7 ENST00000367639.1 |
TEX35
|
testis expressed 35 |
| chr12_+_10010627 | 0.88 |
ENST00000338896.11
ENST00000396502.5 |
CLEC12B
|
C-type lectin domain family 12 member B |
| chr9_+_108862255 | 0.88 |
ENST00000333999.5
|
ACTL7A
|
actin like 7A |
| chr1_-_66801276 | 0.88 |
ENST00000304526.3
|
INSL5
|
insulin like 5 |
| chr11_-_129024157 | 0.87 |
ENST00000392657.7
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr1_+_117001744 | 0.86 |
ENST00000256652.8
ENST00000682167.1 ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr8_-_142777802 | 0.86 |
ENST00000621401.4
|
LYNX1
|
Ly6/neurotoxin 1 |
| chr20_+_4686448 | 0.86 |
ENST00000379440.9
ENST00000424424.2 ENST00000457586.2 |
PRNP
|
prion protein |
| chr15_+_77420668 | 0.85 |
ENST00000381714.7
ENST00000558651.5 |
HMG20A
|
high mobility group 20A |
| chr3_-_20012250 | 0.85 |
ENST00000389050.5
|
PP2D1
|
protein phosphatase 2C like domain containing 1 |
| chr3_+_111674654 | 0.84 |
ENST00000636933.1
ENST00000393934.7 ENST00000477665.2 |
PLCXD2
|
phosphatidylinositol specific phospholipase C X domain containing 2 |
| chr20_+_9514340 | 0.83 |
ENST00000427562.6
|
LAMP5
|
lysosomal associated membrane protein family member 5 |
| chr1_-_173205543 | 0.83 |
ENST00000367718.5
|
TNFSF4
|
TNF superfamily member 4 |
| chr11_+_12094008 | 0.83 |
ENST00000532179.5
ENST00000526065.1 |
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr2_-_206159509 | 0.82 |
ENST00000423725.5
|
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr14_+_101809795 | 0.82 |
ENST00000350249.7
ENST00000557621.5 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2 regulatory subunit B'gamma |
| chr1_+_39158841 | 0.81 |
ENST00000672812.1
|
MACF1
|
microtubule actin crosslinking factor 1 |
| chr1_+_36084079 | 0.80 |
ENST00000207457.8
|
TEKT2
|
tektin 2 |
| chr1_+_177170916 | 0.80 |
ENST00000361539.5
|
BRINP2
|
BMP/retinoic acid inducible neural specific 2 |
| chrX_+_106168297 | 0.80 |
ENST00000337685.6
ENST00000357175.6 |
PWWP3B
|
PWWP domain containing 3B |
| chr20_+_15196834 | 0.79 |
ENST00000402914.5
|
MACROD2
|
mono-ADP ribosylhydrolase 2 |
| chr5_-_140633656 | 0.78 |
ENST00000519715.1
|
CD14
|
CD14 molecule |
| chr13_+_108269629 | 0.78 |
ENST00000430559.5
ENST00000375887.9 |
TNFSF13B
|
TNF superfamily member 13b |
| chr18_+_57352541 | 0.78 |
ENST00000324000.4
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr2_+_230759918 | 0.78 |
ENST00000614925.1
|
CAB39
|
calcium binding protein 39 |
| chr5_+_162068031 | 0.77 |
ENST00000356592.8
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr12_-_10130241 | 0.77 |
ENST00000353231.9
ENST00000525605.1 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr4_-_159035226 | 0.77 |
ENST00000434826.3
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr3_+_141262614 | 0.77 |
ENST00000504264.5
|
PXYLP1
|
2-phosphoxylose phosphatase 1 |
| chr12_+_28452493 | 0.76 |
ENST00000542801.5
|
CCDC91
|
coiled-coil domain containing 91 |
| chr3_-_109118098 | 0.76 |
ENST00000483760.1
|
MORC1
|
MORC family CW-type zinc finger 1 |
| chr17_+_68525795 | 0.75 |
ENST00000592800.5
|
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chr5_+_162067858 | 0.74 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr11_-_105035113 | 0.74 |
ENST00000526568.5
ENST00000531166.5 ENST00000534497.5 ENST00000527979.5 ENST00000533400.6 ENST00000528974.1 ENST00000525825.5 ENST00000353247.9 ENST00000446369.5 ENST00000436863.7 |
CASP1
|
caspase 1 |
| chr1_-_212414815 | 0.72 |
ENST00000261455.9
ENST00000535273.2 |
PACC1
|
proton activated chloride channel 1 |
| chr10_+_89332484 | 0.72 |
ENST00000371811.4
ENST00000680037.1 ENST00000679583.1 ENST00000679897.1 |
IFIT3
|
interferon induced protein with tetratricopeptide repeats 3 |
| chr6_+_101181254 | 0.71 |
ENST00000682090.1
ENST00000421544.6 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr7_+_123601859 | 0.70 |
ENST00000437535.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr5_-_140633690 | 0.70 |
ENST00000512545.1
ENST00000401743.6 |
CD14
|
CD14 molecule |
| chr2_+_11556337 | 0.70 |
ENST00000234142.9
|
GREB1
|
growth regulating estrogen receptor binding 1 |
| chr9_+_128566741 | 0.70 |
ENST00000630866.1
|
SPTAN1
|
spectrin alpha, non-erythrocytic 1 |
| chr4_-_185649524 | 0.69 |
ENST00000451974.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_173635332 | 0.68 |
ENST00000417563.3
|
TEX50
|
testis expressed 50 |
| chr16_-_21652598 | 0.68 |
ENST00000569602.1
ENST00000268389.6 |
IGSF6
|
immunoglobulin superfamily member 6 |
| chr2_-_206159194 | 0.68 |
ENST00000455934.6
ENST00000449699.5 ENST00000454195.1 |
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr5_+_173890545 | 0.68 |
ENST00000519152.5
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr5_-_140633639 | 0.67 |
ENST00000498971.6
|
CD14
|
CD14 molecule |
| chr6_+_101398788 | 0.66 |
ENST00000369138.5
ENST00000413795.5 ENST00000358361.7 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr8_-_33599935 | 0.66 |
ENST00000523956.1
ENST00000256261.9 |
DUSP26
|
dual specificity phosphatase 26 |
| chr17_-_76141240 | 0.65 |
ENST00000322957.7
|
FOXJ1
|
forkhead box J1 |
| chr11_-_84166998 | 0.65 |
ENST00000398299.1
|
DLG2
|
discs large MAGUK scaffold protein 2 |
| chr5_+_58491427 | 0.64 |
ENST00000396776.6
ENST00000502276.6 ENST00000511930.2 |
GAPT
|
GRB2 binding adaptor protein, transmembrane |
| chr19_-_51417700 | 0.64 |
ENST00000529627.1
ENST00000439889.6 |
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chrX_-_6535118 | 0.63 |
ENST00000381089.7
ENST00000612369.4 ENST00000398729.1 |
VCX3A
|
variable charge X-linked 3A |
| chr1_+_103561757 | 0.63 |
ENST00000435302.5
|
AMY2B
|
amylase alpha 2B |
| chr7_-_105269007 | 0.63 |
ENST00000357311.7
|
SRPK2
|
SRSF protein kinase 2 |
| chr5_-_142686047 | 0.63 |
ENST00000360966.9
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 |
| chr5_+_162067990 | 0.63 |
ENST00000641017.1
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr7_+_48924559 | 0.63 |
ENST00000650262.1
|
CDC14C
|
cell division cycle 14C |
| chr8_+_66775178 | 0.63 |
ENST00000396596.2
ENST00000521960.5 ENST00000522398.5 ENST00000522629.5 ENST00000520976.5 |
SGK3
|
serum/glucocorticoid regulated kinase family member 3 |
| chr11_+_5689691 | 0.62 |
ENST00000425490.5
|
TRIM22
|
tripartite motif containing 22 |
| chr17_+_68035636 | 0.62 |
ENST00000676857.1
ENST00000676594.1 ENST00000678388.1 |
KPNA2
|
karyopherin subunit alpha 2 |
| chr5_+_134967901 | 0.62 |
ENST00000282611.8
|
CATSPER3
|
cation channel sperm associated 3 |
| chr19_-_51417791 | 0.60 |
ENST00000353836.9
|
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chrX_-_77969638 | 0.60 |
ENST00000458128.3
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr5_-_11588796 | 0.60 |
ENST00000513598.5
|
CTNND2
|
catenin delta 2 |
| chrY_+_14056226 | 0.59 |
ENST00000250823.5
|
VCY1B
|
variable charge Y-linked 1B |
| chr5_+_169695135 | 0.59 |
ENST00000523684.5
ENST00000519734.5 |
DOCK2
|
dedicator of cytokinesis 2 |
| chr4_+_121801311 | 0.58 |
ENST00000379663.7
ENST00000243498.10 ENST00000509800.5 |
EXOSC9
|
exosome component 9 |
| chr1_+_198638457 | 0.58 |
ENST00000367379.6
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr7_+_123601836 | 0.57 |
ENST00000434204.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr8_-_132675533 | 0.57 |
ENST00000620350.5
ENST00000518642.5 ENST00000250173.5 |
LRRC6
|
leucine rich repeat containing 6 |
| chr9_-_5833014 | 0.57 |
ENST00000339450.10
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr15_+_77420880 | 0.57 |
ENST00000336216.9
ENST00000558176.1 |
HMG20A
|
high mobility group 20A |
| chr20_-_31390580 | 0.57 |
ENST00000339144.3
ENST00000376321.4 |
DEFB119
|
defensin beta 119 |
| chr15_-_42208153 | 0.57 |
ENST00000348544.4
ENST00000318006.10 |
VPS39
|
VPS39 subunit of HOPS complex |
| chr3_-_49429252 | 0.56 |
ENST00000615713.4
|
NICN1
|
nicolin 1 |
| chr5_+_142907573 | 0.55 |
ENST00000469131.6
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr18_+_34710249 | 0.55 |
ENST00000680346.1
ENST00000348997.9 ENST00000681274.1 ENST00000680822.1 ENST00000680767.2 ENST00000597599.5 ENST00000444659.6 |
DTNA
|
dystrobrevin alpha |
| chr14_-_80231052 | 0.55 |
ENST00000557010.5
|
DIO2
|
iodothyronine deiodinase 2 |
| chr5_-_177780633 | 0.54 |
ENST00000513554.5
ENST00000440605.7 |
FAM153A
|
family with sequence similarity 153 member A |
| chr3_-_108058361 | 0.54 |
ENST00000398258.7
|
CD47
|
CD47 molecule |
| chr1_+_174875505 | 0.52 |
ENST00000486220.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr1_-_120051714 | 0.52 |
ENST00000579475.7
|
NOTCH2
|
notch receptor 2 |
| chr12_-_10130082 | 0.52 |
ENST00000533022.5
|
CLEC7A
|
C-type lectin domain containing 7A |
| chr11_+_27055215 | 0.52 |
ENST00000525090.1
|
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr3_+_72888031 | 0.51 |
ENST00000389617.9
|
GXYLT2
|
glucoside xylosyltransferase 2 |
| chr5_+_160009113 | 0.51 |
ENST00000522793.5
ENST00000682719.1 ENST00000684137.1 ENST00000683219.1 ENST00000684018.1 ENST00000231238.10 ENST00000682131.1 |
TTC1
|
tetratricopeptide repeat domain 1 |
| chr1_+_86547070 | 0.50 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr14_+_103385374 | 0.50 |
ENST00000678179.1
ENST00000676938.1 ENST00000678619.1 ENST00000440884.7 ENST00000560417.6 ENST00000679330.1 ENST00000556744.2 ENST00000676897.1 ENST00000677560.1 ENST00000561314.6 ENST00000677829.1 ENST00000677133.1 ENST00000676645.1 ENST00000678175.1 ENST00000429436.7 ENST00000677360.1 ENST00000678237.1 ENST00000677347.1 ENST00000677432.1 |
MARK3
|
microtubule affinity regulating kinase 3 |
| chr3_-_49429304 | 0.50 |
ENST00000636166.1
ENST00000273598.8 ENST00000436744.2 |
ENSG00000283189.2
NICN1
|
novel protein nicolin 1 |
| chr8_+_27380629 | 0.49 |
ENST00000412793.5
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr17_-_62806632 | 0.49 |
ENST00000583803.1
ENST00000456609.6 |
MARCHF10
|
membrane associated ring-CH-type finger 10 |
| chr1_+_78649818 | 0.48 |
ENST00000370747.9
ENST00000438486.1 |
IFI44
|
interferon induced protein 44 |
| chr11_-_26572254 | 0.48 |
ENST00000529533.6
|
MUC15
|
mucin 15, cell surface associated |
| chr8_-_53842899 | 0.48 |
ENST00000524234.1
ENST00000521275.5 ENST00000396774.6 |
ATP6V1H
|
ATPase H+ transporting V1 subunit H |
| chr2_-_169694332 | 0.48 |
ENST00000421028.1
|
CCDC173
|
coiled-coil domain containing 173 |
| chr15_+_21579912 | 0.48 |
ENST00000628444.1
|
LINC02203
|
long intergenic non-protein coding RNA 2203 |
| chr3_+_131026844 | 0.48 |
ENST00000510769.5
ENST00000383366.9 ENST00000510688.5 ENST00000511262.5 |
NEK11
|
NIMA related kinase 11 |
| chrX_+_136648214 | 0.47 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr10_-_114144599 | 0.46 |
ENST00000428953.1
|
CCDC186
|
coiled-coil domain containing 186 |
| chr15_+_22015233 | 0.46 |
ENST00000639059.1
ENST00000640156.1 |
ENSG00000285472.1
ENSG00000284500.3
|
novel protein novel transcript |
| chr1_-_17045219 | 0.46 |
ENST00000491274.5
|
SDHB
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr4_-_99435396 | 0.46 |
ENST00000209665.8
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr2_-_112433519 | 0.45 |
ENST00000496537.1
ENST00000330575.9 ENST00000302558.8 |
RGPD8
|
RANBP2 like and GRIP domain containing 8 |
| chr7_+_142791635 | 0.45 |
ENST00000633705.1
|
TRBC1
|
T cell receptor beta constant 1 |
| chr7_+_123601815 | 0.45 |
ENST00000451215.6
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr1_-_155969724 | 0.45 |
ENST00000609707.1
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr12_+_29149238 | 0.45 |
ENST00000536681.8
|
FAR2
|
fatty acyl-CoA reductase 2 |
| chr16_+_18983927 | 0.45 |
ENST00000569532.5
ENST00000304381.10 |
TMC7
|
transmembrane channel like 7 |
| chr2_-_73779968 | 0.45 |
ENST00000452812.1
ENST00000443070.5 ENST00000272444.7 |
DUSP11
|
dual specificity phosphatase 11 |
| chr4_+_94974984 | 0.45 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr1_-_170074568 | 0.44 |
ENST00000367767.5
ENST00000361580.7 ENST00000538366.5 |
KIFAP3
|
kinesin associated protein 3 |
| chr3_-_196270540 | 0.44 |
ENST00000419333.5
|
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr16_+_31873772 | 0.44 |
ENST00000394846.7
ENST00000300870.15 |
ZNF267
|
zinc finger protein 267 |
| chr2_-_98663464 | 0.43 |
ENST00000414521.6
|
MGAT4A
|
alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A |
| chr2_+_109794296 | 0.43 |
ENST00000430736.5
ENST00000016946.8 ENST00000441344.1 |
RGPD5
|
RANBP2 like and GRIP domain containing 5 |
| chrX_+_136648138 | 0.43 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand |
| chr16_-_11281322 | 0.43 |
ENST00000312511.4
|
PRM1
|
protamine 1 |
| chr4_-_175891691 | 0.43 |
ENST00000507540.1
|
GPM6A
|
glycoprotein M6A |
| chr12_+_9827517 | 0.43 |
ENST00000537723.5
|
KLRF1
|
killer cell lectin like receptor F1 |
| chr11_-_46119787 | 0.42 |
ENST00000529782.5
ENST00000532010.5 ENST00000525438.5 ENST00000533757.5 ENST00000527782.5 |
PHF21A
|
PHD finger protein 21A |
| chr8_+_103880412 | 0.41 |
ENST00000436393.6
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chrX_+_77910656 | 0.41 |
ENST00000343533.9
ENST00000341514.11 ENST00000645454.1 ENST00000642651.1 ENST00000644362.1 |
ATP7A
PGK1
|
ATPase copper transporting alpha phosphoglycerate kinase 1 |
| chr12_-_86256267 | 0.41 |
ENST00000620241.4
|
MGAT4C
|
MGAT4 family member C |
| chr10_-_30629741 | 0.41 |
ENST00000647634.2
ENST00000375318.4 |
LYZL2
|
lysozyme like 2 |
| chr4_+_168654724 | 0.40 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr17_-_73227700 | 0.40 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104 member A |
| chr10_-_24622817 | 0.39 |
ENST00000636789.1
|
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr3_-_131026726 | 0.39 |
ENST00000514044.5
ENST00000264992.8 |
ASTE1
|
asteroid homolog 1 |
| chr4_+_94207596 | 0.39 |
ENST00000359052.8
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr12_+_74537787 | 0.39 |
ENST00000519948.4
|
ATXN7L3B
|
ataxin 7 like 3B |
| chr12_-_49130113 | 0.39 |
ENST00000549870.5
|
TUBA1B
|
tubulin alpha 1b |
| chrX_+_23908006 | 0.39 |
ENST00000379211.8
ENST00000648352.1 |
CXorf58
|
chromosome X open reading frame 58 |
| chr6_+_32439866 | 0.39 |
ENST00000374982.5
ENST00000395388.7 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr4_-_99435336 | 0.39 |
ENST00000437033.7
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr1_-_60073750 | 0.39 |
ENST00000371201.3
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr12_-_118359639 | 0.38 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr12_+_125065427 | 0.38 |
ENST00000316519.11
|
AACS
|
acetoacetyl-CoA synthetase |
| chr2_+_11542662 | 0.38 |
ENST00000389825.7
ENST00000381483.6 |
GREB1
|
growth regulating estrogen receptor binding 1 |
| chr3_+_122055355 | 0.38 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chr22_-_39893755 | 0.37 |
ENST00000325157.7
|
ENTHD1
|
ENTH domain containing 1 |
| chr2_-_110577101 | 0.37 |
ENST00000330331.9
ENST00000446930.1 ENST00000329516.8 |
RGPD6
|
RANBP2 like and GRIP domain containing 6 |
| chr6_+_110180418 | 0.37 |
ENST00000368930.5
ENST00000307731.2 |
CDC40
|
cell division cycle 40 |
| chr13_+_48256214 | 0.37 |
ENST00000650237.1
|
ITM2B
|
integral membrane protein 2B |
| chr12_-_68159732 | 0.36 |
ENST00000229135.4
|
IFNG
|
interferon gamma |
Network of associatons between targets according to the STRING database.
First level regulatory network of EMX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.4 | 1.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.3 | 3.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.3 | 0.9 | GO:0002769 | natural killer cell inhibitory signaling pathway(GO:0002769) |
| 0.3 | 1.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.3 | 1.5 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.3 | 1.7 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.3 | 0.8 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.3 | 0.8 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.3 | 2.2 | GO:0071725 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.2 | 0.7 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 0.9 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.2 | 0.8 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.2 | 0.8 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.2 | 0.6 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.2 | 6.9 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 0.7 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.2 | 1.0 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.2 | 1.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.4 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.1 | 1.8 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.4 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 1.8 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.1 | 1.4 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.4 | GO:0060003 | copper ion export(GO:0060003) |
| 0.1 | 1.4 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.1 | 0.4 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.8 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.1 | 0.4 | GO:0002912 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.1 | 0.4 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) positive regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000309) |
| 0.1 | 0.4 | GO:0060723 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.1 | 0.6 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.1 | 4.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.6 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.6 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.1 | 2.3 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.3 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.2 | GO:0016121 | tetraterpenoid metabolic process(GO:0016108) carotenoid metabolic process(GO:0016116) carotene catabolic process(GO:0016121) xanthophyll metabolic process(GO:0016122) terpene catabolic process(GO:0046247) |
| 0.1 | 0.5 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 0.6 | GO:0072139 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 | 0.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.6 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.6 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 1.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.8 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 3.0 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 2.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 1.9 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 1.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 2.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.9 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.2 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.3 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.1 | 0.2 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 0.3 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.1 | 1.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 1.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 2.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.5 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.6 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.5 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.3 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 0.8 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.8 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 0.7 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.5 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.6 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 1.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 2.2 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.4 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.9 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 1.2 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.2 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.2 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.0 | 0.2 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.6 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 1.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.4 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.6 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 2.2 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 1.5 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.4 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 2.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 1.7 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.9 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.9 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.4 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 1.2 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 3.5 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.7 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 1.1 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 3.6 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.4 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.5 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.0 | 0.3 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.4 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.3 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.2 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.6 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.1 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0046070 | dGTP metabolic process(GO:0046070) |
| 0.0 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.3 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.8 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 2.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.0 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 1.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.2 | 2.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 2.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 2.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 1.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.4 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 3.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.7 | GO:0072558 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 1.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.3 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.1 | 0.5 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.5 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.5 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 1.8 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 4.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.4 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.9 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 2.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.6 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 2.6 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.6 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 1.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 1.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.4 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 2.9 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 2.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 3.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 2.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 2.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.4 | 1.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.3 | 1.7 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.3 | 0.9 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.3 | 1.2 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.3 | 2.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.3 | 2.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 1.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.2 | 0.9 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.2 | 3.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.4 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.1 | 1.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.4 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) superoxide dismutase copper chaperone activity(GO:0016532) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 0.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.6 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 3.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.6 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 1.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.5 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 2.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.5 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.6 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.9 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 0.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 2.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.3 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.1 | 2.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.3 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 0.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 8.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.9 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 2.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.9 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.1 | 4.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.9 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.4 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 0.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 2.3 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.3 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 2.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.5 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.0 | 1.0 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 2.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 1.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 1.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 1.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.4 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0004320 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.0 | 2.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.5 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 1.6 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 2.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 1.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 2.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 4.2 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 3.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.5 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 3.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.8 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |