Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
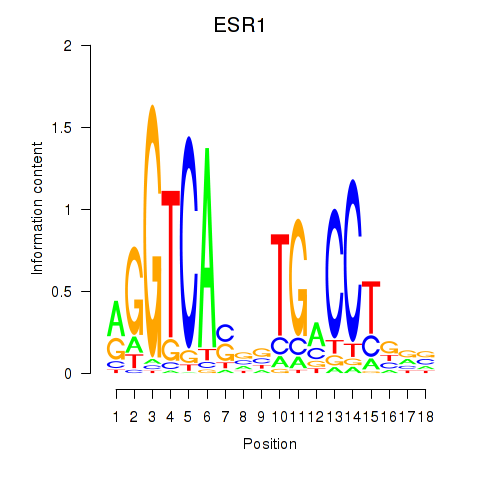
Results for ESR1
Z-value: 2.49
Transcription factors associated with ESR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESR1
|
ENSG00000091831.24 | estrogen receptor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ESR1 | hg38_v1_chr6_+_151809105_151809139 | 0.25 | 1.6e-01 | Click! |
Activity profile of ESR1 motif
Sorted Z-values of ESR1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_22686724 | 19.35 |
ENST00000390305.2
|
IGLV3-25
|
immunoglobulin lambda variable 3-25 |
| chr22_+_22758698 | 15.61 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr22_+_22409755 | 10.19 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr22_+_22792485 | 9.91 |
ENST00000390314.2
|
IGLV2-11
|
immunoglobulin lambda variable 2-11 |
| chr22_+_22369601 | 9.81 |
ENST00000390295.3
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 |
| chr14_-_105588322 | 9.54 |
ENST00000497872.4
ENST00000390539.2 |
IGHA2
|
immunoglobulin heavy constant alpha 2 (A2m marker) |
| chr22_+_22880706 | 9.26 |
ENST00000390319.2
|
IGLV3-1
|
immunoglobulin lambda variable 3-1 |
| chr2_-_88947820 | 9.11 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr22_+_22380766 | 9.00 |
ENST00000390297.3
|
IGLV1-44
|
immunoglobulin lambda variable 1-44 |
| chr2_-_89100352 | 8.88 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr22_+_22195753 | 8.80 |
ENST00000390285.4
|
IGLV6-57
|
immunoglobulin lambda variable 6-57 |
| chr2_-_89010515 | 7.98 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr22_+_22322452 | 7.88 |
ENST00000390290.3
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr22_+_22357739 | 7.87 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr2_-_89213917 | 7.81 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr2_-_89268506 | 7.67 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chr22_+_22906342 | 7.27 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr2_-_89117844 | 7.06 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr22_+_22395005 | 6.98 |
ENST00000390298.2
|
IGLV7-43
|
immunoglobulin lambda variable 7-43 |
| chr2_-_88966767 | 6.93 |
ENST00000464162.1
|
IGKV1-6
|
immunoglobulin kappa variable 1-6 |
| chr22_+_22697789 | 6.50 |
ENST00000390306.2
|
IGLV2-23
|
immunoglobulin lambda variable 2-23 |
| chr22_+_22895368 | 6.49 |
ENST00000390321.2
|
IGLC1
|
immunoglobulin lambda constant 1 |
| chr14_-_105708627 | 6.40 |
ENST00000641837.1
ENST00000390547.3 |
IGHA1
|
immunoglobulin heavy constant alpha 1 |
| chr2_-_88992903 | 6.29 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr22_+_22822658 | 6.12 |
ENST00000620395.2
|
IGLV2-8
|
immunoglobulin lambda variable 2-8 |
| chr22_+_22900976 | 5.96 |
ENST00000390323.2
|
IGLC2
|
immunoglobulin lambda constant 2 |
| chr2_+_89862438 | 5.87 |
ENST00000448155.2
|
IGKV1D-39
|
immunoglobulin kappa variable 1D-39 |
| chr14_-_106360320 | 5.78 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
| chr14_-_106269133 | 5.67 |
ENST00000390609.3
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr14_-_106762576 | 5.65 |
ENST00000624687.1
|
IGHV1-69D
|
immunoglobulin heavy variable 1-69D |
| chr22_+_22030934 | 5.52 |
ENST00000390282.2
|
IGLV4-69
|
immunoglobulin lambda variable 4-69 |
| chr8_+_11494367 | 5.28 |
ENST00000259089.9
ENST00000529894.1 |
BLK
|
BLK proto-oncogene, Src family tyrosine kinase |
| chr2_-_88979016 | 5.21 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr2_+_90154073 | 5.12 |
ENST00000611391.1
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr14_-_106579223 | 5.07 |
ENST00000390626.2
|
IGHV5-51
|
immunoglobulin heavy variable 5-51 |
| chr22_+_22327298 | 4.97 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr22_+_22668286 | 4.93 |
ENST00000390304.2
|
IGLV3-27
|
immunoglobulin lambda variable 3-27 |
| chr17_-_35880350 | 4.85 |
ENST00000605140.6
ENST00000651122.1 ENST00000603197.6 |
CCL5
|
C-C motif chemokine ligand 5 |
| chr19_-_7732160 | 4.75 |
ENST00000676742.1
ENST00000678118.1 ENST00000328853.10 |
CLEC4G
|
C-type lectin domain family 4 member G |
| chr22_+_22343185 | 4.69 |
ENST00000427632.2
|
IGLV9-49
|
immunoglobulin lambda variable 9-49 |
| chr17_+_36064265 | 4.50 |
ENST00000616054.2
|
CCL18
|
C-C motif chemokine ligand 18 |
| chr14_-_106639589 | 4.42 |
ENST00000390630.3
|
IGHV4-61
|
immunoglobulin heavy variable 4-61 |
| chr17_-_3691748 | 4.31 |
ENST00000552723.1
|
P2RX5
|
purinergic receptor P2X 5 |
| chr14_-_94390667 | 4.22 |
ENST00000557492.5
ENST00000355814.8 ENST00000437397.5 ENST00000448921.5 ENST00000393088.8 |
SERPINA1
|
serpin family A member 1 |
| chr19_-_10334723 | 4.13 |
ENST00000592945.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr2_+_230225718 | 4.10 |
ENST00000420434.7
ENST00000392045.8 ENST00000417495.7 ENST00000343805.10 |
SP140
|
SP140 nuclear body protein |
| chr15_-_22160868 | 3.98 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr19_+_49335396 | 3.97 |
ENST00000598095.5
ENST00000426897.6 ENST00000323906.9 ENST00000535669.6 ENST00000597602.1 ENST00000595660.1 |
CD37
|
CD37 molecule |
| chr14_-_94390650 | 3.97 |
ENST00000449399.7
ENST00000404814.8 |
SERPINA1
|
serpin family A member 1 |
| chr15_+_32718476 | 3.94 |
ENST00000652365.1
|
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr2_+_90038848 | 3.90 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr14_-_94390614 | 3.83 |
ENST00000553327.5
ENST00000556955.5 ENST00000557118.5 ENST00000440909.5 |
SERPINA1
|
serpin family A member 1 |
| chr1_+_206897435 | 3.74 |
ENST00000391929.7
ENST00000294984.7 ENST00000611909.4 ENST00000367093.3 |
IL24
|
interleukin 24 |
| chr14_-_106005574 | 3.48 |
ENST00000390595.3
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr22_+_22098683 | 3.46 |
ENST00000390283.2
|
IGLV8-61
|
immunoglobulin lambda variable 8-61 |
| chr11_+_67404077 | 3.35 |
ENST00000542590.2
ENST00000312390.9 |
TBC1D10C
|
TBC1 domain family member 10C |
| chr2_-_89297785 | 3.33 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr6_-_2903300 | 3.13 |
ENST00000380698.5
|
SERPINB9
|
serpin family B member 9 |
| chr22_+_22904850 | 3.06 |
ENST00000390324.2
|
IGLJ3
|
immunoglobulin lambda joining 3 |
| chr22_-_23754376 | 3.03 |
ENST00000398465.3
ENST00000248948.4 |
VPREB3
|
V-set pre-B cell surrogate light chain 3 |
| chr16_+_30183595 | 2.97 |
ENST00000219150.10
ENST00000570045.5 ENST00000565497.5 ENST00000570244.5 |
CORO1A
|
coronin 1A |
| chr14_+_100019375 | 2.87 |
ENST00000544450.6
|
EVL
|
Enah/Vasp-like |
| chr9_-_120926752 | 2.85 |
ENST00000373887.8
|
TRAF1
|
TNF receptor associated factor 1 |
| chr6_-_30686624 | 2.84 |
ENST00000274853.8
|
PPP1R18
|
protein phosphatase 1 regulatory subunit 18 |
| chr19_+_544034 | 2.82 |
ENST00000592501.5
ENST00000264553.6 |
GZMM
|
granzyme M |
| chr2_+_118942188 | 2.80 |
ENST00000327097.5
|
MARCO
|
macrophage receptor with collagenous structure |
| chr3_+_46979659 | 2.75 |
ENST00000450053.8
|
NBEAL2
|
neurobeachin like 2 |
| chr3_+_186930518 | 2.75 |
ENST00000169298.8
ENST00000457772.6 ENST00000455441.5 ENST00000427315.5 |
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr15_+_88639009 | 2.67 |
ENST00000306072.10
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr3_+_186930759 | 2.66 |
ENST00000677292.1
ENST00000458216.5 |
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr5_-_67196791 | 2.63 |
ENST00000256447.5
|
CD180
|
CD180 molecule |
| chr19_+_18173804 | 2.62 |
ENST00000407280.4
|
IFI30
|
IFI30 lysosomal thiol reductase |
| chr4_-_154590735 | 2.61 |
ENST00000403106.8
ENST00000622532.1 ENST00000651975.1 |
FGA
|
fibrinogen alpha chain |
| chr11_+_67403887 | 2.57 |
ENST00000526387.5
|
TBC1D10C
|
TBC1 domain family member 10C |
| chr1_+_161707244 | 2.55 |
ENST00000349527.8
ENST00000294796.8 ENST00000309691.10 ENST00000367953.7 ENST00000367950.2 |
FCRLA
|
Fc receptor like A |
| chr2_+_230225756 | 2.54 |
ENST00000373645.3
|
SP140
|
SP140 nuclear body protein |
| chr6_+_37005630 | 2.51 |
ENST00000274963.13
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr2_+_89985922 | 2.47 |
ENST00000390268.2
|
IGKV2D-26
|
immunoglobulin kappa variable 2D-26 |
| chr6_+_35297809 | 2.44 |
ENST00000316637.7
|
DEF6
|
DEF6 guanine nucleotide exchange factor |
| chr19_-_7747511 | 2.41 |
ENST00000593660.5
ENST00000204801.12 ENST00000315591.12 ENST00000354397.10 ENST00000394161.9 ENST00000593821.5 ENST00000602261.5 ENST00000601256.1 ENST00000601951.5 ENST00000315599.12 |
CD209
|
CD209 molecule |
| chr15_+_81196871 | 2.40 |
ENST00000559383.5
ENST00000394660.6 ENST00000683961.1 |
IL16
|
interleukin 16 |
| chr17_+_50634845 | 2.38 |
ENST00000427699.5
ENST00000285238.13 |
ABCC3
|
ATP binding cassette subfamily C member 3 |
| chr6_-_73452253 | 2.35 |
ENST00000370318.5
ENST00000370315.4 |
CGAS
|
cyclic GMP-AMP synthase |
| chr3_-_183555696 | 2.34 |
ENST00000341319.8
|
KLHL6
|
kelch like family member 6 |
| chr12_-_14843517 | 2.33 |
ENST00000228936.6
|
ART4
|
ADP-ribosyltransferase 4 (inactive) (Dombrock blood group) |
| chr19_-_3606849 | 2.32 |
ENST00000375190.10
|
TBXA2R
|
thromboxane A2 receptor |
| chr15_+_80933358 | 2.22 |
ENST00000560027.1
|
CEMIP
|
cell migration inducing hyaluronidase 1 |
| chr6_+_31494881 | 2.19 |
ENST00000538442.5
|
MICB
|
MHC class I polypeptide-related sequence B |
| chr2_+_218323148 | 2.19 |
ENST00000258362.7
|
PNKD
|
PNKD metallo-beta-lactamase domain containing |
| chr15_+_88638947 | 2.16 |
ENST00000559876.2
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr19_+_50415799 | 2.16 |
ENST00000599632.1
|
ENSG00000142539.9
|
novel protein |
| chr17_-_31318818 | 2.09 |
ENST00000578584.5
|
ENSG00000265118.5
|
novel protein |
| chr5_+_40679907 | 2.08 |
ENST00000302472.4
|
PTGER4
|
prostaglandin E receptor 4 |
| chr19_-_4540028 | 2.07 |
ENST00000306390.7
|
LRG1
|
leucine rich alpha-2-glycoprotein 1 |
| chr1_-_111427731 | 2.07 |
ENST00000369732.4
|
OVGP1
|
oviductal glycoprotein 1 |
| chr7_+_2632029 | 2.05 |
ENST00000407643.5
|
TTYH3
|
tweety family member 3 |
| chr11_+_67289283 | 2.02 |
ENST00000511455.7
|
ANKRD13D
|
ankyrin repeat domain 13D |
| chr15_+_32717994 | 1.97 |
ENST00000560677.5
ENST00000560830.1 ENST00000651154.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr22_+_22747383 | 1.96 |
ENST00000390311.3
|
IGLV3-16
|
immunoglobulin lambda variable 3-16 |
| chr1_+_9651723 | 1.96 |
ENST00000377346.9
ENST00000536656.5 ENST00000628140.2 |
PIK3CD
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr1_+_196774813 | 1.96 |
ENST00000471440.6
ENST00000391985.7 ENST00000617219.1 ENST00000367425.9 |
CFHR3
|
complement factor H related 3 |
| chr5_-_140633167 | 1.95 |
ENST00000302014.11
|
CD14
|
CD14 molecule |
| chr11_+_47248885 | 1.94 |
ENST00000395397.7
ENST00000405576.5 |
NR1H3
|
nuclear receptor subfamily 1 group H member 3 |
| chr20_+_63736651 | 1.93 |
ENST00000487026.5
ENST00000309546.8 ENST00000480139.5 |
LIME1
|
Lck interacting transmembrane adaptor 1 |
| chr1_+_161707222 | 1.92 |
ENST00000236938.12
|
FCRLA
|
Fc receptor like A |
| chr22_+_39456996 | 1.92 |
ENST00000341184.7
|
MGAT3
|
beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase |
| chr8_-_21913671 | 1.91 |
ENST00000523932.1
ENST00000276420.9 |
DOK2
|
docking protein 2 |
| chr2_+_118942290 | 1.87 |
ENST00000412481.1
|
MARCO
|
macrophage receptor with collagenous structure |
| chr3_-_50303565 | 1.86 |
ENST00000266031.8
ENST00000395143.6 ENST00000457214.6 ENST00000447605.2 ENST00000395144.7 ENST00000418723.1 |
HYAL1
|
hyaluronidase 1 |
| chr1_+_26021768 | 1.86 |
ENST00000374280.4
|
EXTL1
|
exostosin like glycosyltransferase 1 |
| chr16_-_74700845 | 1.85 |
ENST00000308807.12
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain like pseudokinase |
| chr19_+_17281863 | 1.83 |
ENST00000652132.1
ENST00000404085.7 ENST00000598347.2 |
ANKLE1
|
ankyrin repeat and LEM domain containing 1 |
| chr1_+_37474572 | 1.82 |
ENST00000373087.7
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr1_-_206772484 | 1.82 |
ENST00000423557.1
|
IL10
|
interleukin 10 |
| chr14_+_52314280 | 1.77 |
ENST00000557436.1
ENST00000245457.6 |
PTGER2
|
prostaglandin E receptor 2 |
| chr16_+_1240698 | 1.76 |
ENST00000561736.2
ENST00000338844.8 ENST00000461509.6 |
TPSAB1
|
tryptase alpha/beta 1 |
| chr19_+_7355589 | 1.76 |
ENST00000671891.2
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor 18 |
| chr11_+_1870150 | 1.76 |
ENST00000429923.5
ENST00000418975.1 ENST00000406638.6 |
LSP1
|
lymphocyte specific protein 1 |
| chr16_+_29459913 | 1.74 |
ENST00000360423.12
|
SULT1A4
|
sulfotransferase family 1A member 4 |
| chr16_-_74700786 | 1.72 |
ENST00000306247.11
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain like pseudokinase |
| chr19_-_3606577 | 1.72 |
ENST00000411851.3
|
TBXA2R
|
thromboxane A2 receptor |
| chr6_-_30687200 | 1.71 |
ENST00000399199.7
|
PPP1R18
|
protein phosphatase 1 regulatory subunit 18 |
| chr11_+_1870252 | 1.69 |
ENST00000612798.4
|
LSP1
|
lymphocyte specific protein 1 |
| chr15_-_72783685 | 1.69 |
ENST00000456471.3
ENST00000311669.12 |
ADPGK
|
ADP dependent glucokinase |
| chr14_-_91253925 | 1.68 |
ENST00000531499.2
|
GPR68
|
G protein-coupled receptor 68 |
| chr21_-_44914271 | 1.68 |
ENST00000522931.5
|
ITGB2
|
integrin subunit beta 2 |
| chr11_+_102317450 | 1.67 |
ENST00000615299.4
ENST00000527309.2 ENST00000526421.6 ENST00000263464.9 |
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr6_-_73452124 | 1.67 |
ENST00000680833.1
|
CGAS
|
cyclic GMP-AMP synthase |
| chr22_+_22899481 | 1.66 |
ENST00000390322.2
|
IGLJ2
|
immunoglobulin lambda joining 2 |
| chr19_-_7747559 | 1.65 |
ENST00000394173.8
|
CD209
|
CD209 molecule |
| chr6_-_41039202 | 1.62 |
ENST00000244565.8
|
UNC5CL
|
unc-5 family C-terminal like |
| chr16_+_726936 | 1.62 |
ENST00000549114.5
ENST00000341413.8 ENST00000562187.1 ENST00000564537.5 ENST00000389703.8 |
HAGHL
|
hydroxyacylglutathione hydrolase like |
| chr7_+_99374240 | 1.61 |
ENST00000443222.6
ENST00000414376.5 |
ARPC1B
|
actin related protein 2/3 complex subunit 1B |
| chr7_+_45574358 | 1.61 |
ENST00000297323.12
|
ADCY1
|
adenylate cyclase 1 |
| chr3_-_46464868 | 1.61 |
ENST00000417439.5
ENST00000231751.9 ENST00000431944.1 |
LTF
|
lactotransferrin |
| chr16_-_1230089 | 1.59 |
ENST00000612142.1
ENST00000606293.5 |
TPSB2
|
tryptase beta 2 |
| chr11_+_47248924 | 1.58 |
ENST00000481889.6
ENST00000436778.5 ENST00000531660.5 ENST00000407404.5 |
NR1H3
|
nuclear receptor subfamily 1 group H member 3 |
| chr6_-_149484965 | 1.58 |
ENST00000409806.8
|
ZC3H12D
|
zinc finger CCCH-type containing 12D |
| chr16_+_30199255 | 1.57 |
ENST00000338971.10
|
SULT1A3
|
sulfotransferase family 1A member 3 |
| chr9_-_121370275 | 1.57 |
ENST00000538954.5
|
STOM
|
stomatin |
| chr11_+_65639860 | 1.57 |
ENST00000527525.5
|
SIPA1
|
signal-induced proliferation-associated 1 |
| chr11_+_102317542 | 1.56 |
ENST00000532808.5
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr11_+_36296281 | 1.54 |
ENST00000530639.6
|
PRR5L
|
proline rich 5 like |
| chr8_-_21913661 | 1.53 |
ENST00000518197.1
|
DOK2
|
docking protein 2 |
| chr8_-_69833338 | 1.51 |
ENST00000524945.5
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1 |
| chr7_+_99374675 | 1.51 |
ENST00000645391.1
ENST00000455009.6 |
ARPC1B
|
actin related protein 2/3 complex subunit 1B |
| chr1_+_32362537 | 1.50 |
ENST00000373534.4
|
TSSK3
|
testis specific serine kinase 3 |
| chr9_+_133534807 | 1.50 |
ENST00000393060.1
|
ADAMTSL2
|
ADAMTS like 2 |
| chr3_+_122055355 | 1.49 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chr7_-_1940492 | 1.47 |
ENST00000437877.1
|
MAD1L1
|
mitotic arrest deficient 1 like 1 |
| chr11_+_102317492 | 1.47 |
ENST00000673846.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr6_-_90296824 | 1.47 |
ENST00000257749.9
|
BACH2
|
BTB domain and CNC homolog 2 |
| chr7_+_2631978 | 1.46 |
ENST00000258796.12
|
TTYH3
|
tweety family member 3 |
| chr2_-_207166818 | 1.46 |
ENST00000423015.5
|
KLF7
|
Kruppel like factor 7 |
| chr22_+_22588155 | 1.45 |
ENST00000390302.3
|
IGLV2-33
|
immunoglobulin lambda variable 2-33 (non-functional) |
| chr9_+_133534697 | 1.44 |
ENST00000651351.2
|
ADAMTSL2
|
ADAMTS like 2 |
| chr6_+_29827817 | 1.43 |
ENST00000360323.11
ENST00000376818.7 ENST00000376815.3 |
HLA-G
|
major histocompatibility complex, class I, G |
| chr9_-_35619542 | 1.43 |
ENST00000396757.6
|
CD72
|
CD72 molecule |
| chr16_+_30199860 | 1.43 |
ENST00000395138.6
|
SULT1A3
|
sulfotransferase family 1A member 3 |
| chr1_-_19980416 | 1.41 |
ENST00000375111.7
|
PLA2G2A
|
phospholipase A2 group IIA |
| chr19_-_54313074 | 1.40 |
ENST00000486742.2
ENST00000432233.8 |
LILRA5
|
leukocyte immunoglobulin like receptor A5 |
| chr22_-_50270353 | 1.40 |
ENST00000330651.11
|
MAPK11
|
mitogen-activated protein kinase 11 |
| chr20_+_62656359 | 1.39 |
ENST00000370507.5
|
SLCO4A1
|
solute carrier organic anion transporter family member 4A1 |
| chr8_+_141128581 | 1.39 |
ENST00000519811.6
|
DENND3
|
DENN domain containing 3 |
| chr2_-_98731063 | 1.37 |
ENST00000393487.6
|
MGAT4A
|
alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A |
| chr21_-_44928711 | 1.37 |
ENST00000517563.5
|
ITGB2
|
integrin subunit beta 2 |
| chr20_+_38346474 | 1.36 |
ENST00000217407.3
|
LBP
|
lipopolysaccharide binding protein |
| chr10_+_87659839 | 1.36 |
ENST00000456849.2
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr1_-_156020789 | 1.35 |
ENST00000531917.5
ENST00000480567.5 ENST00000526212.2 ENST00000529008.5 ENST00000496742.5 ENST00000295702.9 |
SSR2
|
signal sequence receptor subunit 2 |
| chr7_-_150341615 | 1.33 |
ENST00000223271.8
ENST00000466675.5 ENST00000482669.1 ENST00000467793.5 |
RARRES2
|
retinoic acid receptor responder 2 |
| chr7_+_142492121 | 1.32 |
ENST00000390374.3
|
TRBV7-6
|
T cell receptor beta variable 7-6 |
| chr19_+_45499610 | 1.30 |
ENST00000396735.6
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent 1N (putative) |
| chr6_-_90296908 | 1.30 |
ENST00000537989.5
|
BACH2
|
BTB domain and CNC homolog 2 |
| chr8_+_143558329 | 1.30 |
ENST00000262580.9
ENST00000525721.1 ENST00000534018.5 |
GSDMD
|
gasdermin D |
| chr5_+_111224374 | 1.28 |
ENST00000282356.9
|
CAMK4
|
calcium/calmodulin dependent protein kinase IV |
| chr8_+_141128612 | 1.27 |
ENST00000518347.5
ENST00000262585.6 ENST00000520986.5 ENST00000523058.5 ENST00000518668.5 |
DENND3
|
DENN domain containing 3 |
| chr11_+_62419025 | 1.26 |
ENST00000278282.3
|
SCGB1A1
|
secretoglobin family 1A member 1 |
| chr11_+_8683201 | 1.24 |
ENST00000526562.5
ENST00000525981.1 |
RPL27A
|
ribosomal protein L27a |
| chr6_-_53348902 | 1.24 |
ENST00000370913.5
ENST00000304434.11 |
ELOVL5
|
ELOVL fatty acid elongase 5 |
| chr13_-_46390042 | 1.23 |
ENST00000389908.7
|
RUBCNL
|
rubicon like autophagy enhancer |
| chr19_-_13102848 | 1.23 |
ENST00000264824.5
|
LYL1
|
LYL1 basic helix-loop-helix family member |
| chr4_+_2798564 | 1.23 |
ENST00000504294.5
|
SH3BP2
|
SH3 domain binding protein 2 |
| chr22_+_37639660 | 1.22 |
ENST00000649765.2
ENST00000451997.6 |
SH3BP1
ENSG00000285304.1
|
SH3 domain binding protein 1 novel protein |
| chr19_+_17281645 | 1.22 |
ENST00000394458.7
ENST00000594072.6 |
ANKLE1
|
ankyrin repeat and LEM domain containing 1 |
| chr11_+_8682782 | 1.21 |
ENST00000531978.5
ENST00000524496.5 ENST00000532359.5 ENST00000314138.11 ENST00000530022.5 |
RPL27A
|
ribosomal protein L27a |
| chr20_+_33993932 | 1.21 |
ENST00000333552.9
|
RALY
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr7_-_100573865 | 1.20 |
ENST00000622764.3
|
SAP25
|
Sin3A associated protein 25 |
| chr9_+_127716073 | 1.20 |
ENST00000373289.4
|
TTC16
|
tetratricopeptide repeat domain 16 |
| chr1_+_12166978 | 1.19 |
ENST00000376259.7
ENST00000536782.2 |
TNFRSF1B
|
TNF receptor superfamily member 1B |
| chr19_-_14475307 | 1.19 |
ENST00000292513.4
|
PTGER1
|
prostaglandin E receptor 1 |
| chr22_+_49918733 | 1.18 |
ENST00000407217.7
ENST00000403427.3 |
CRELD2
|
cysteine rich with EGF like domains 2 |
| chr17_-_75405486 | 1.18 |
ENST00000392562.5
|
GRB2
|
growth factor receptor bound protein 2 |
| chr5_+_111223905 | 1.18 |
ENST00000512453.5
|
CAMK4
|
calcium/calmodulin dependent protein kinase IV |
| chr6_+_32154131 | 1.18 |
ENST00000375143.6
ENST00000324816.11 ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr19_+_47332167 | 1.17 |
ENST00000595464.3
|
C5AR2
|
complement component 5a receptor 2 |
| chr14_+_21768482 | 1.17 |
ENST00000390428.3
|
TRAV6
|
T cell receptor alpha variable 6 |
| chr5_-_132011580 | 1.17 |
ENST00000651250.1
ENST00000434099.6 ENST00000296869.9 ENST00000651356.1 ENST00000651883.2 |
ACSL6
|
acyl-CoA synthetase long chain family member 6 |
| chr3_+_186931344 | 1.17 |
ENST00000417392.5
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr15_+_69452811 | 1.17 |
ENST00000357790.5
ENST00000260379.11 ENST00000560274.1 |
RPLP1
|
ribosomal protein lateral stalk subunit P1 |
| chr5_-_132011811 | 1.16 |
ENST00000379255.5
ENST00000430403.5 ENST00000357096.5 |
ACSL6
|
acyl-CoA synthetase long chain family member 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ESR1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 1.6 | 4.9 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 1.6 | 17.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 1.2 | 208.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 1.2 | 4.8 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.2 | 4.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.0 | 3.1 | GO:0002372 | myeloid dendritic cell cytokine production(GO:0002372) |
| 1.0 | 3.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 1.0 | 4.1 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.8 | 2.4 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.7 | 3.5 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.7 | 2.1 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.6 | 1.8 | GO:2000627 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.6 | 1.8 | GO:0060302 | negative regulation of interleukin-18 production(GO:0032701) negative regulation of cytokine activity(GO:0060302) negative regulation of chemokine (C-C motif) ligand 5 production(GO:0071650) |
| 0.6 | 1.8 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.6 | 3.0 | GO:0032796 | uropod organization(GO:0032796) |
| 0.6 | 2.3 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.5 | 5.9 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.5 | 2.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.5 | 0.5 | GO:0002458 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) |
| 0.5 | 1.5 | GO:0043017 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.5 | 1.4 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.4 | 2.6 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.4 | 1.3 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.4 | 3.0 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.4 | 2.9 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.4 | 1.6 | GO:0044010 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.4 | 2.4 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.4 | 2.8 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.4 | 1.6 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.4 | 3.0 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.4 | 1.1 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.3 | 1.0 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.3 | 1.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.3 | 2.6 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.3 | 1.0 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.3 | 70.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.3 | 2.2 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.3 | 1.9 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.3 | 0.9 | GO:0006212 | uracil catabolic process(GO:0006212) beta-alanine biosynthetic process(GO:0019483) |
| 0.3 | 1.7 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.3 | 1.4 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.3 | 0.8 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.3 | 7.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.3 | 0.8 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.3 | 2.4 | GO:0034127 | regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034127) negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.3 | 1.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.3 | 0.8 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.3 | 2.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.3 | 1.3 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.3 | 1.3 | GO:0010193 | response to ozone(GO:0010193) |
| 0.2 | 0.7 | GO:1903450 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.2 | 1.0 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.2 | 4.7 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.2 | 1.1 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.2 | 0.7 | GO:1904863 | regulation of beta-catenin-TCF complex assembly(GO:1904863) negative regulation of beta-catenin-TCF complex assembly(GO:1904864) |
| 0.2 | 2.9 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 1.3 | GO:0019732 | antifungal humoral response(GO:0019732) antifungal innate immune response(GO:0061760) |
| 0.2 | 0.9 | GO:0051801 | cytolysis in other organism involved in symbiotic interaction(GO:0051801) |
| 0.2 | 4.8 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.2 | 2.2 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.2 | 1.0 | GO:0038156 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.2 | 0.6 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.2 | 1.0 | GO:0071603 | retinal blood vessel morphogenesis(GO:0061304) endothelial cell-cell adhesion(GO:0071603) |
| 0.2 | 2.9 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 0.6 | GO:1904887 | regulation of thioredoxin peroxidase activity(GO:1903123) negative regulation of thioredoxin peroxidase activity(GO:1903124) negative regulation of thioredoxin peroxidase activity by peptidyl-threonine phosphorylation(GO:1903125) Wnt signalosome assembly(GO:1904887) negative regulation of peroxidase activity(GO:2000469) |
| 0.2 | 2.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 0.6 | GO:0060003 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) copper ion export(GO:0060003) |
| 0.2 | 0.5 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.2 | 0.5 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 0.7 | GO:0033590 | acetaldehyde metabolic process(GO:0006117) response to cobalamin(GO:0033590) |
| 0.2 | 0.7 | GO:0014028 | notochord formation(GO:0014028) |
| 0.2 | 0.7 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.2 | 1.7 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.2 | 1.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.2 | 1.4 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 1.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 0.6 | GO:0060734 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.2 | 0.5 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 0.9 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.6 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.1 | 11.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 1.3 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.1 | 0.9 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.1 | 5.2 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 3.2 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 3.9 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.1 | 0.7 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 1.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.4 | GO:0097212 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.1 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.7 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 4.9 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 | 1.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 5.3 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 | 0.5 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.1 | 0.8 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 | 0.8 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.1 | 0.2 | GO:0051885 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of anagen(GO:0051885) |
| 0.1 | 0.5 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.1 | 0.4 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.1 | 0.9 | GO:0072343 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.5 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 1.5 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.1 | 1.3 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 0.3 | GO:1903988 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.1 | 2.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.7 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 | 1.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 4.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 2.3 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 1.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.1 | 0.6 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.1 | 0.6 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 0.7 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.1 | 1.8 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 0.6 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.5 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 2.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 1.9 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.1 | 0.7 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.4 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.1 | 4.0 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.4 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 | 0.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 4.8 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 1.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 2.4 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.1 | 2.7 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 0.5 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 0.4 | GO:0000050 | urea cycle(GO:0000050) |
| 0.1 | 2.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.3 | GO:0070895 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.1 | 0.3 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.1 | 0.4 | GO:1900220 | negative regulation of alkaline phosphatase activity(GO:0010693) semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.6 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 1.7 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 1.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 1.6 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 3.0 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 0.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.2 | GO:0048561 | establishment of organ orientation(GO:0048561) |
| 0.1 | 1.5 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 0.7 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 0.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 1.6 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 0.4 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 1.0 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 2.3 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.1 | 0.4 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 21.8 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 0.8 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 0.1 | 1.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.8 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.1 | 0.4 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) |
| 0.1 | 0.5 | GO:0061430 | bone trabecula morphogenesis(GO:0061430) |
| 0.1 | 0.5 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 0.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.5 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.6 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 2.3 | GO:0050779 | RNA destabilization(GO:0050779) |
| 0.1 | 0.8 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.1 | 0.6 | GO:0032682 | negative regulation of chemokine production(GO:0032682) |
| 0.1 | 0.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.2 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 0.3 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.7 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 2.1 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.7 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.6 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.8 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.3 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 1.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.3 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.5 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.6 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 3.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.8 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.4 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.7 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 2.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 1.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.2 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 4.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.3 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.4 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.6 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 4.6 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.5 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.9 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.3 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.6 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.2 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 1.4 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 0.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.2 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 1.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 2.5 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.3 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.9 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.4 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 2.6 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.5 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 2.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.5 | GO:0001659 | temperature homeostasis(GO:0001659) |
| 0.0 | 0.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.5 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.0 | GO:0032077 | positive regulation of deoxyribonuclease activity(GO:0032077) |
| 0.0 | 0.8 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 1.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.5 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.3 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.9 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.0 | GO:1903626 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 | 0.9 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 1.1 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.4 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.2 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.5 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.2 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) |
| 0.0 | 0.2 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 15.9 | GO:0071752 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) |
| 1.0 | 3.1 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.7 | 2.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.7 | 35.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.6 | 1.9 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.4 | 1.9 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.4 | 3.0 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.4 | 1.5 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.3 | 73.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 1.0 | GO:0030526 | granulocyte macrophage colony-stimulating factor receptor complex(GO:0030526) |
| 0.3 | 1.6 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.2 | 7.7 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.2 | 2.8 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 1.0 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 1.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 0.6 | GO:0099400 | cytoplasmic side of mitochondrial outer membrane(GO:0032473) caveola neck(GO:0099400) |
| 0.2 | 1.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 0.7 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.1 | 1.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.8 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 1.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.8 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 1.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.8 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.4 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 12.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 6.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 1.3 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 2.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.6 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.1 | 7.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.9 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.3 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.1 | 1.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.3 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 1.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.4 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 1.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 5.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 6.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 2.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.2 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 5.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.4 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 84.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 8.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 4.1 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 1.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 1.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 5.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 3.1 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 3.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 2.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.8 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 1.8 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 95.1 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 4.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.0 | 0.5 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.8 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 1.6 | 4.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.1 | 303.7 | GO:0003823 | antigen binding(GO:0003823) |
| 1.0 | 4.8 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.8 | 2.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.8 | 2.4 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.8 | 2.3 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.7 | 3.0 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.7 | 6.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.6 | 1.8 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.5 | 3.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.5 | 2.9 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.5 | 6.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.5 | 1.9 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.4 | 3.5 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.4 | 4.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.4 | 1.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.4 | 3.3 | GO:0071723 | lipoteichoic acid binding(GO:0070891) lipopeptide binding(GO:0071723) |
| 0.4 | 4.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.4 | 1.4 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.3 | 1.7 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.3 | 3.0 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.3 | 2.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.3 | 0.8 | GO:0090422 | thiamine pyrophosphate transporter activity(GO:0090422) |
| 0.3 | 1.1 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.2 | 0.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 2.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 2.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 0.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 1.0 | GO:0004914 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 1.7 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 1.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.2 | 0.6 | GO:0036479 | GTP-dependent protein kinase activity(GO:0034211) peroxidase inhibitor activity(GO:0036479) |
| 0.2 | 0.6 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 0.9 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.2 | 0.5 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.2 | 1.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 2.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 0.7 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.2 | 0.7 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.2 | 1.0 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.2 | 0.6 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.2 | 3.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 3.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 1.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.6 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 2.8 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 3.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 4.7 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 1.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 2.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 1.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 3.9 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.8 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.7 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 1.0 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 7.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 3.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.6 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 1.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 2.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 3.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.7 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.1 | 3.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 4.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.3 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.1 | 2.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.5 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.1 | 3.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.8 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 0.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.5 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.5 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 1.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.0 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 0.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.4 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.6 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 0.7 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.8 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 1.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.2 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 12.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 1.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.9 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.1 | 0.3 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.1 | 0.8 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 1.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.8 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.5 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 1.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.4 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 1.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.5 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.5 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 1.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 5.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.7 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.3 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.9 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.5 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 1.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 1.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 1.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0030290 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.0 | 0.7 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.4 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 1.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 1.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 2.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 1.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.6 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.5 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.6 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 7.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 5.5 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 5.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.9 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 2.0 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.1 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.0 | 0.1 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.0 | 1.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 4.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 4.3 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.3 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 1.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.0 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.6 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.1 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.8 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 3.7 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.8 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 3.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 2.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 9.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 12.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 6.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 4.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 4.4 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 3.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 4.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 3.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 5.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 3.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 0.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 4.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 9.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 1.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 3.9 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 5.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 3.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 6.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 6.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.0 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 9.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 9.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.3 | 2.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.2 | 3.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 10.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 3.6 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 3.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 2.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 4.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 6.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.5 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 9.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 4.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 10.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 3.6 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 2.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 4.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.9 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 1.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 5.1 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 3.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 5.5 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 2.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 2.5 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 1.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 2.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |