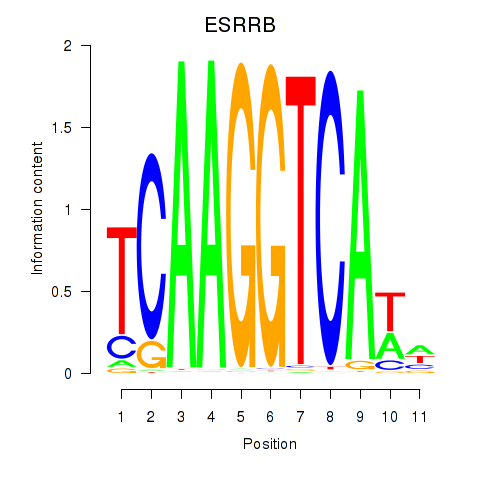
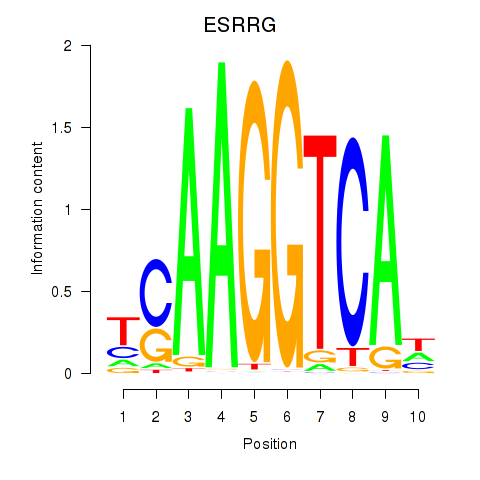
Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for ESRRB_ESRRG
Z-value: 3.34
Transcription factors associated with ESRRB_ESRRG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESRRB
|
ENSG00000119715.15 | estrogen related receptor beta |
|
ESRRG
|
ENSG00000196482.18 | estrogen related receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ESRRG | hg38_v1_chr1_-_216723437_216723472 | 0.83 | 4.7e-09 | Click! |
| ESRRB | hg38_v1_chr14_+_76376113_76376151 | 0.52 | 2.1e-03 | Click! |
Activity profile of ESRRB_ESRRG motif
Sorted Z-values of ESRRB_ESRRG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ESRRB_ESRRG
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.0 | 63.9 | GO:0060262 | regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 9.8 | 29.4 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 4.1 | 12.4 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 3.1 | 12.5 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 2.7 | 45.8 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 2.6 | 15.6 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 2.4 | 16.5 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 2.3 | 9.3 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 2.2 | 2.2 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 2.0 | 54.6 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 1.9 | 20.8 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 1.9 | 5.7 | GO:0042323 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 1.8 | 14.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 1.8 | 3.5 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 1.7 | 5.2 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 1.7 | 11.7 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 1.7 | 8.3 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 1.6 | 30.6 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.4 | 5.7 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 1.4 | 24.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 1.4 | 5.7 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 1.2 | 7.2 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 1.2 | 3.6 | GO:2000296 | regulation of hydrogen peroxide catabolic process(GO:2000295) negative regulation of hydrogen peroxide catabolic process(GO:2000296) regulation of oxygen metabolic process(GO:2000374) |
| 1.2 | 3.6 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 1.2 | 20.0 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 1.1 | 3.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 1.1 | 19.0 | GO:0042407 | cristae formation(GO:0042407) |
| 1.1 | 3.2 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 1.1 | 3.2 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 1.1 | 3.2 | GO:0080033 | cellular response to nitrite(GO:0071250) response to nitrite(GO:0080033) positive regulation of mitochondrial DNA metabolic process(GO:1901860) glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) response to resveratrol(GO:1904638) cellular response to resveratrol(GO:1904639) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 1.0 | 4.2 | GO:0021893 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.9 | 3.8 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.9 | 5.4 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.9 | 2.6 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.8 | 4.1 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.8 | 2.4 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.8 | 3.2 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 0.8 | 2.4 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.8 | 3.9 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.8 | 3.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.8 | 12.1 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.7 | 3.0 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.7 | 4.4 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.7 | 4.1 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.7 | 5.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.6 | 0.6 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.6 | 45.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.6 | 2.6 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.6 | 1.9 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.6 | 8.6 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.6 | 3.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.6 | 3.7 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.6 | 1.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.5 | 6.8 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.5 | 3.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.5 | 4.2 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.5 | 7.7 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.5 | 2.0 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.5 | 4.6 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.5 | 5.0 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.5 | 13.0 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.5 | 11.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.5 | 12.9 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.5 | 5.7 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.5 | 0.9 | GO:0034759 | detoxification of copper ion(GO:0010273) regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) stress response to copper ion(GO:1990169) |
| 0.4 | 1.8 | GO:1903610 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 0.4 | 5.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.4 | 1.2 | GO:0071650 | negative regulation of chemokine (C-C motif) ligand 5 production(GO:0071650) |
| 0.4 | 2.4 | GO:1901908 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.4 | 6.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.4 | 0.8 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.4 | 4.7 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.4 | 3.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.4 | 12.9 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.4 | 1.9 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.4 | 1.8 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.4 | 1.1 | GO:0051939 | gamma-aminobutyric acid import(GO:0051939) |
| 0.3 | 1.7 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.3 | 10.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.3 | 2.4 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.3 | 3.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 2.3 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.3 | 5.5 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.3 | 9.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 4.9 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 8.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.3 | 1.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.3 | 8.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.3 | 3.0 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.3 | 2.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.3 | 1.2 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.3 | 3.5 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.3 | 5.7 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.3 | 9.1 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.3 | 1.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.3 | 4.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.3 | 4.6 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.3 | 1.3 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.3 | 1.6 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.3 | 6.8 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.3 | 5.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.3 | 9.6 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.3 | 1.3 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.3 | 5.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 3.8 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.3 | 5.8 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 22.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 4.1 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.2 | 7.0 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.2 | 6.9 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.2 | 0.9 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 | 4.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 3.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.2 | 0.7 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.2 | 3.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 4.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 1.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.5 | GO:0018032 | protein amidation(GO:0018032) |
| 0.2 | 1.6 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.2 | 4.7 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.2 | 4.1 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.2 | 0.6 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 2.9 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 0.9 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.2 | 1.5 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 8.6 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.2 | 0.6 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.2 | 1.3 | GO:1903294 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 0.2 | 4.8 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.2 | 2.0 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 1.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 1.4 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.2 | 4.0 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 3.1 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.2 | 16.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 2.2 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.2 | 2.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.2 | 2.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.2 | 0.6 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.2 | 1.8 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.2 | 1.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 1.0 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.2 | 10.3 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.2 | 3.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.2 | 0.5 | GO:0035572 | N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.2 | 1.5 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.2 | 2.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 1.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 2.3 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.2 | 4.1 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 8.3 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 1.6 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 3.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 3.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 3.5 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.1 | 3.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.4 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.1 | 6.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.7 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.1 | 4.3 | GO:0070875 | positive regulation of glycogen biosynthetic process(GO:0045725) positive regulation of glycogen metabolic process(GO:0070875) |
| 0.1 | 0.5 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.1 | 5.5 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 1.6 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 2.8 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 0.1 | 1.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 1.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 3.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 2.9 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 1.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 2.1 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 1.7 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 2.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 1.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 4.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 17.3 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 6.1 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 2.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 1.8 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.1 | 3.7 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.4 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 4.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.5 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 3.9 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 0.3 | GO:1904875 | regulation of DNA ligase activity(GO:1904875) |
| 0.1 | 1.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.8 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 3.6 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.1 | 0.9 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.8 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 2.1 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.1 | 1.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 5.1 | GO:0001662 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.1 | 0.9 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 1.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 1.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 3.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.1 | GO:0097017 | renal protein absorption(GO:0097017) |
| 0.1 | 0.9 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 4.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 3.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.7 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 1.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 1.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 12.3 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.1 | 7.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 1.6 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 1.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.8 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.8 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 1.6 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 3.4 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 2.8 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 1.6 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 7.2 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.1 | 1.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 8.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.8 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.1 | 7.1 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 2.1 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 0.3 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 1.0 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.6 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 1.9 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 0.7 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 1.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 9.5 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.1 | 1.7 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 2.0 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 2.7 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.1 | 2.0 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 3.2 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 1.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 1.5 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 3.5 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 0.7 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.1 | 4.5 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.1 | 1.4 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 7.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 2.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 2.8 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 0.6 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.8 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) regulation of protein complex stability(GO:0061635) |
| 0.1 | 2.3 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 2.4 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 | 1.7 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 2.1 | GO:0021695 | cerebellar cortex development(GO:0021695) |
| 0.0 | 9.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 5.5 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 0.7 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 | 0.7 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.8 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 1.4 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 1.6 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 2.2 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 1.0 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.2 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.2 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 1.9 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 2.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 1.8 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 0.0 | 0.6 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.0 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 1.2 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 4.8 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 2.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.4 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 2.5 | GO:0046034 | ATP metabolic process(GO:0046034) |
| 0.0 | 1.3 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.0 | 3.9 | GO:0008652 | cellular amino acid biosynthetic process(GO:0008652) |
| 0.0 | 0.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 1.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 3.6 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.4 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.6 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.0 | 5.7 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.3 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.3 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.5 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 2.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.2 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 1.4 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 1.2 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.3 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.8 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.4 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.0 | 0.1 | GO:0097286 | iron ion import(GO:0097286) |
| 0.0 | 0.9 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.7 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.8 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 4.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.9 | GO:0000045 | autophagosome assembly(GO:0000045) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 54.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 2.9 | 11.7 | GO:0005745 | m-AAA complex(GO:0005745) |
| 2.6 | 44.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 2.4 | 14.4 | GO:0070470 | plasma membrane respiratory chain(GO:0070470) |
| 2.2 | 8.6 | GO:0098855 | HCN channel complex(GO:0098855) |
| 1.5 | 24.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 1.2 | 4.7 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 1.1 | 9.6 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 1.1 | 3.2 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 1.0 | 9.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.0 | 6.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.9 | 5.5 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.9 | 33.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.9 | 4.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.7 | 1.4 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.7 | 1.4 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.7 | 2.6 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.6 | 15.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.6 | 5.4 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.6 | 5.9 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.6 | 10.5 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.5 | 3.7 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.5 | 7.7 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.5 | 2.0 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.5 | 52.2 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.4 | 1.3 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.4 | 7.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.4 | 3.9 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.4 | 2.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.4 | 5.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.4 | 3.7 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.4 | 4.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.4 | 3.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.4 | 23.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 21.8 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 3.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 5.8 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.3 | 3.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.3 | 1.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 7.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 3.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 10.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 8.2 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 0.6 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.2 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 12.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 3.6 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.2 | 12.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 4.2 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 4.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.0 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.2 | 12.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 37.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 19.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 3.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 1.9 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 5.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 6.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 2.2 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 0.8 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 3.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.9 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 3.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 7.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 3.1 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 3.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 2.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 8.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.4 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 1.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 3.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 8.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 25.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.3 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.1 | 1.8 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 4.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 5.5 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 7.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 44.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 0.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 1.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.3 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 1.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 22.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 11.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.2 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.1 | 0.8 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 6.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 8.0 | GO:0030016 | myofibril(GO:0030016) |
| 0.1 | 4.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 3.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.3 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 2.4 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 56.5 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 21.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.6 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 13.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 10.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 12.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 1.5 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 1.3 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 6.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 5.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 3.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 3.5 | GO:0019866 | organelle inner membrane(GO:0019866) |
| 0.0 | 2.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 2.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.8 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 7.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.9 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 5.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 6.3 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 10.4 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 3.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 9.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.9 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 45.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 4.9 | 29.4 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 3.1 | 9.3 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 2.6 | 7.8 | GO:0080130 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 2.2 | 13.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 1.9 | 7.6 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 1.5 | 56.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 1.4 | 7.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 1.4 | 13.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 1.4 | 5.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 1.1 | 5.7 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 1.1 | 5.7 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 1.1 | 8.6 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.0 | 9.4 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 1.0 | 4.0 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.8 | 5.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.8 | 4.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.8 | 19.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.8 | 4.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.8 | 4.0 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.8 | 3.9 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.8 | 5.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.8 | 26.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.7 | 2.2 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.7 | 20.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.7 | 4.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.6 | 3.2 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.6 | 12.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.6 | 3.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.6 | 1.8 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.6 | 8.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.6 | 2.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.6 | 5.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.6 | 34.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.6 | 7.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.5 | 2.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.5 | 1.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.5 | 3.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.5 | 2.4 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.5 | 1.9 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.5 | 2.9 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.5 | 1.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.5 | 4.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.4 | 1.8 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 0.4 | 1.3 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.4 | 4.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.4 | 3.9 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.4 | 8.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 2.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.4 | 7.0 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.4 | 2.4 | GO:0052845 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.4 | 3.9 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.4 | 3.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.4 | 3.7 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.4 | 12.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 1.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.3 | 6.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 3.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.3 | 6.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.3 | 4.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 10.9 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.3 | 8.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 15.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 0.9 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.3 | 3.7 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.3 | 2.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.3 | 1.2 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.3 | 2.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.3 | 2.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 2.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.3 | 2.7 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.3 | 1.9 | GO:0046624 | sphingolipid transporter activity(GO:0046624) ceramide binding(GO:0097001) |
| 0.3 | 3.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.3 | 2.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 3.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 4.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 1.3 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.3 | 3.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.3 | 6.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 2.0 | GO:0035473 | lipase binding(GO:0035473) |
| 0.2 | 1.2 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 3.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 1.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 1.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 1.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 3.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 1.8 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 3.6 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.2 | 2.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 5.8 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 5.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 2.6 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.2 | 3.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 0.6 | GO:0046715 | borate transmembrane transporter activity(GO:0046715) |
| 0.2 | 1.5 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.2 | 3.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 1.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 7.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 4.2 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.2 | 0.6 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.2 | 5.0 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 6.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 7.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 0.7 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.2 | 3.9 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 4.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.2 | 3.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 2.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 2.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.6 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 8.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 5.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 0.6 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 26.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.8 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.5 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.1 | 3.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 5.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 3.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 7.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.6 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 14.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.7 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.5 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 2.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 1.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 2.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 1.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 1.0 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 4.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.6 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 4.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 3.3 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.9 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 4.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 11.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 4.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.7 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 5.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 2.7 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.0 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 0.4 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 24.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.7 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.6 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.1 | 0.3 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 1.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 2.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 2.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.6 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 0.6 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 1.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.2 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.1 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 1.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.8 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.4 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.8 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 1.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 18.9 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 3.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 6.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 2.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 2.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 3.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 2.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 1.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 1.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.2 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.7 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 2.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 1.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 1.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 3.1 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 2.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 3.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 1.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 2.1 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 3.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.5 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 1.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 1.0 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.8 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 1.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 2.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 3.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 10.9 | GO:0019787 | ubiquitin-like protein transferase activity(GO:0019787) |
| 0.0 | 2.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 12.3 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 2.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 1.5 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 19.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 7.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 8.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 18.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 28.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 21.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 6.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 4.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 6.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 5.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 3.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 10.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 1.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 2.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 1.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 4.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 8.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 6.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 4.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 4.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 5.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 2.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 75.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.9 | 81.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.6 | 15.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.5 | 14.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.4 | 8.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.4 | 26.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 2.9 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.3 | 10.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 4.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.3 | 16.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 11.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 8.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 4.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 11.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 2.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 7.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 31.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 51.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.2 | 8.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 2.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 5.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 4.7 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 3.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 4.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 3.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 3.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 3.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.6 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.1 | 3.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 13.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.8 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 18.3 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 4.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.7 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 13.0 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 1.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 3.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 1.8 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 2.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 2.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 4.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 10.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 2.4 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 2.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 3.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |