Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
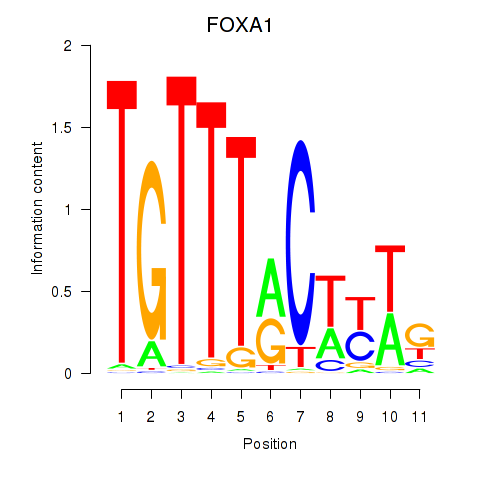
Results for FOXA1
Z-value: 1.81
Transcription factors associated with FOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA1
|
ENSG00000129514.8 | forkhead box A1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA1 | hg38_v1_chr14_-_37595224_37595281 | 0.63 | 1.1e-04 | Click! |
Activity profile of FOXA1 motif
Sorted Z-values of FOXA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_154612635 | 10.68 |
ENST00000407946.5
ENST00000405164.5 ENST00000336098.8 ENST00000393846.6 ENST00000404648.7 ENST00000443553.5 |
FGG
|
fibrinogen gamma chain |
| chr17_-_28368012 | 10.45 |
ENST00000555059.2
|
ENSG00000273171.1
|
novel protein, readthrough between VTN and SEBOX |
| chr10_-_79560386 | 9.99 |
ENST00000372327.9
ENST00000417041.1 ENST00000640627.1 ENST00000372325.7 |
SFTPA2
|
surfactant protein A2 |
| chr10_+_79610932 | 9.43 |
ENST00000428376.6
ENST00000398636.8 ENST00000419470.6 ENST00000429958.5 |
SFTPA1
|
surfactant protein A1 |
| chr11_-_6440980 | 8.35 |
ENST00000265983.8
ENST00000615166.1 |
HPX
|
hemopexin |
| chr11_+_62419025 | 8.05 |
ENST00000278282.3
|
SCGB1A1
|
secretoglobin family 1A member 1 |
| chr1_-_56966133 | 7.37 |
ENST00000535057.5
ENST00000543257.5 |
C8B
|
complement C8 beta chain |
| chr14_-_94323324 | 7.24 |
ENST00000341584.4
|
SERPINA6
|
serpin family A member 6 |
| chr1_-_56966006 | 6.82 |
ENST00000371237.9
|
C8B
|
complement C8 beta chain |
| chr17_-_41528293 | 6.20 |
ENST00000455635.1
ENST00000361566.7 |
KRT19
|
keratin 19 |
| chr5_-_41213505 | 6.03 |
ENST00000337836.10
ENST00000433294.1 |
C6
|
complement C6 |
| chr3_+_138010143 | 5.44 |
ENST00000183605.10
|
CLDN18
|
claudin 18 |
| chr4_-_69495897 | 5.27 |
ENST00000305107.7
ENST00000639621.1 |
UGT2B4
|
UDP glucuronosyltransferase family 2 member B4 |
| chr2_-_21044063 | 5.14 |
ENST00000233242.5
|
APOB
|
apolipoprotein B |
| chr11_-_116792386 | 5.14 |
ENST00000433069.2
ENST00000542499.5 |
APOA5
|
apolipoprotein A5 |
| chr10_-_46046264 | 5.09 |
ENST00000581478.5
ENST00000582163.3 |
MSMB
|
microseminoprotein beta |
| chr3_+_137998735 | 5.07 |
ENST00000343735.8
|
CLDN18
|
claudin 18 |
| chr2_-_68952880 | 4.91 |
ENST00000481498.1
ENST00000328895.9 |
GKN2
|
gastrokine 2 |
| chr3_+_186613052 | 4.76 |
ENST00000411641.7
ENST00000273784.5 |
AHSG
|
alpha 2-HS glycoprotein |
| chr4_-_69495861 | 4.69 |
ENST00000512583.5
|
UGT2B4
|
UDP glucuronosyltransferase family 2 member B4 |
| chr22_+_20774092 | 4.68 |
ENST00000215727.10
|
SERPIND1
|
serpin family D member 1 |
| chr3_+_52777580 | 4.65 |
ENST00000273283.7
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr14_-_94323389 | 4.60 |
ENST00000557225.1
|
SERPINA6
|
serpin family A member 6 |
| chr14_-_37595224 | 4.58 |
ENST00000250448.5
|
FOXA1
|
forkhead box A1 |
| chr12_-_52949818 | 4.46 |
ENST00000546897.5
ENST00000552551.5 |
KRT8
|
keratin 8 |
| chr10_+_122560639 | 4.27 |
ENST00000344338.7
ENST00000330163.8 ENST00000652446.2 ENST00000666315.1 ENST00000368955.7 ENST00000368909.7 ENST00000368956.6 ENST00000619379.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr5_-_41213021 | 4.27 |
ENST00000417809.1
|
C6
|
complement C6 |
| chr19_+_18386150 | 4.17 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr17_-_47957824 | 4.11 |
ENST00000300557.3
|
PRR15L
|
proline rich 15 like |
| chr4_+_186227501 | 4.00 |
ENST00000446598.6
ENST00000264690.11 ENST00000513864.2 |
KLKB1
|
kallikrein B1 |
| chr11_-_124752247 | 3.90 |
ENST00000326621.10
|
VSIG2
|
V-set and immunoglobulin domain containing 2 |
| chr8_+_119067239 | 3.87 |
ENST00000332843.3
|
COLEC10
|
collectin subfamily member 10 |
| chr7_-_81770039 | 3.84 |
ENST00000222390.11
ENST00000453411.6 ENST00000457544.7 ENST00000444829.7 |
HGF
|
hepatocyte growth factor |
| chr20_-_7940444 | 3.83 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1 |
| chr4_+_186266183 | 3.75 |
ENST00000403665.7
ENST00000492972.6 ENST00000264692.8 |
F11
|
coagulation factor XI |
| chr10_+_122560751 | 3.71 |
ENST00000338354.10
ENST00000664692.1 ENST00000653442.1 ENST00000664974.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr3_+_186996444 | 3.67 |
ENST00000676633.1
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr12_-_52949776 | 3.59 |
ENST00000548998.5
|
KRT8
|
keratin 8 |
| chr20_+_33007695 | 3.50 |
ENST00000170150.4
|
BPIFB2
|
BPI fold containing family B member 2 |
| chr3_+_186974957 | 3.48 |
ENST00000438590.5
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr21_-_42315336 | 3.40 |
ENST00000398431.2
ENST00000518498.3 |
TFF3
|
trefoil factor 3 |
| chr1_-_26913964 | 3.27 |
ENST00000254227.4
|
NR0B2
|
nuclear receptor subfamily 0 group B member 2 |
| chr2_-_157488829 | 3.24 |
ENST00000435117.1
ENST00000439355.5 |
CYTIP
|
cytohesin 1 interacting protein |
| chr12_-_89352395 | 3.23 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr3_+_52211442 | 3.23 |
ENST00000459884.1
|
ALAS1
|
5'-aminolevulinate synthase 1 |
| chr12_-_52949849 | 3.21 |
ENST00000619952.2
ENST00000546826.5 |
KRT8
|
keratin 8 |
| chr11_-_124752187 | 3.20 |
ENST00000403470.1
|
VSIG2
|
V-set and immunoglobulin domain containing 2 |
| chr3_-_149221811 | 3.16 |
ENST00000455472.3
ENST00000264613.11 |
CP
|
ceruloplasmin |
| chr12_-_89352487 | 3.15 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr13_-_46142834 | 3.14 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr17_+_9021501 | 3.11 |
ENST00000173229.7
|
NTN1
|
netrin 1 |
| chr20_+_33235987 | 3.07 |
ENST00000375422.6
ENST00000375413.8 ENST00000354297.9 |
BPIFA1
|
BPI fold containing family A member 1 |
| chr8_+_125430333 | 3.07 |
ENST00000311922.4
|
TRIB1
|
tribbles pseudokinase 1 |
| chr7_+_73828160 | 2.99 |
ENST00000431918.1
|
CLDN4
|
claudin 4 |
| chr6_+_160702238 | 2.98 |
ENST00000366924.6
ENST00000308192.14 ENST00000418964.1 |
PLG
|
plasminogen |
| chr6_-_41747390 | 2.95 |
ENST00000356667.8
ENST00000373025.7 ENST00000425343.6 |
PGC
|
progastricsin |
| chr10_-_79949098 | 2.81 |
ENST00000372292.8
|
SFTPD
|
surfactant protein D |
| chr14_-_36519679 | 2.80 |
ENST00000498187.6
|
NKX2-1
|
NK2 homeobox 1 |
| chr10_+_122560679 | 2.78 |
ENST00000657942.1
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr4_+_54229261 | 2.64 |
ENST00000508170.5
ENST00000512143.1 ENST00000257290.10 |
PDGFRA
|
platelet derived growth factor receptor alpha |
| chr17_-_50468871 | 2.60 |
ENST00000508540.6
ENST00000258969.4 |
CHAD
|
chondroadherin |
| chr13_+_77535681 | 2.60 |
ENST00000349847.4
|
SCEL
|
sciellin |
| chr13_+_77535742 | 2.59 |
ENST00000377246.7
|
SCEL
|
sciellin |
| chr6_-_46954922 | 2.58 |
ENST00000265417.7
|
ADGRF5
|
adhesion G protein-coupled receptor F5 |
| chr12_-_120327762 | 2.58 |
ENST00000308366.9
ENST00000423423.3 |
PLA2G1B
|
phospholipase A2 group IB |
| chr13_+_77535669 | 2.57 |
ENST00000535157.5
|
SCEL
|
sciellin |
| chr9_+_27109135 | 2.54 |
ENST00000519097.5
ENST00000615002.4 |
TEK
|
TEK receptor tyrosine kinase |
| chr7_+_80602150 | 2.52 |
ENST00000309881.11
|
CD36
|
CD36 molecule |
| chr15_-_55249029 | 2.51 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr7_+_134891400 | 2.47 |
ENST00000393118.6
|
CALD1
|
caldesmon 1 |
| chr7_-_81770013 | 2.45 |
ENST00000465234.2
|
HGF
|
hepatocyte growth factor |
| chr2_-_21043941 | 2.42 |
ENST00000399256.4
|
APOB
|
apolipoprotein B |
| chr12_-_91178520 | 2.42 |
ENST00000425043.5
ENST00000420120.6 ENST00000441303.6 ENST00000456569.2 |
DCN
|
decorin |
| chr7_+_80602200 | 2.42 |
ENST00000534394.5
|
CD36
|
CD36 molecule |
| chrX_-_15384402 | 2.41 |
ENST00000297904.4
|
VEGFD
|
vascular endothelial growth factor D |
| chr18_+_3449620 | 2.38 |
ENST00000405385.7
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr9_+_27109200 | 2.38 |
ENST00000380036.10
|
TEK
|
TEK receptor tyrosine kinase |
| chr1_+_200027702 | 2.37 |
ENST00000367362.8
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2 |
| chr11_-_10568650 | 2.35 |
ENST00000256178.8
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr19_+_18382163 | 2.28 |
ENST00000595973.2
|
GDF15
|
growth differentiation factor 15 |
| chr13_-_48413105 | 2.28 |
ENST00000620633.5
|
LPAR6
|
lysophosphatidic acid receptor 6 |
| chr11_-_108593738 | 2.19 |
ENST00000525344.5
ENST00000265843.9 |
EXPH5
|
exophilin 5 |
| chr3_+_142596385 | 2.15 |
ENST00000457734.7
ENST00000483373.5 ENST00000475296.5 ENST00000495744.5 ENST00000476044.5 ENST00000461644.5 ENST00000464320.5 |
PLS1
|
plastin 1 |
| chr7_-_81770122 | 2.15 |
ENST00000423064.7
|
HGF
|
hepatocyte growth factor |
| chr14_-_69797232 | 2.12 |
ENST00000216540.5
|
SLC10A1
|
solute carrier family 10 member 1 |
| chr3_+_148730100 | 2.11 |
ENST00000474935.5
ENST00000475347.5 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor type 1 |
| chr1_+_200027605 | 2.09 |
ENST00000236914.7
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2 |
| chr19_-_48615050 | 2.06 |
ENST00000263266.4
|
FAM83E
|
family with sequence similarity 83 member E |
| chr13_-_85799400 | 2.04 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr2_-_88861563 | 1.98 |
ENST00000624935.3
ENST00000390241.3 |
ENSG00000240040.6
IGKJ2
|
novel transcript immunoglobulin kappa joining 2 |
| chr8_-_132760548 | 1.96 |
ENST00000519187.5
ENST00000523829.5 ENST00000677595.1 ENST00000356838.7 ENST00000377901.8 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr7_+_135148041 | 1.95 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr5_+_102865737 | 1.94 |
ENST00000509523.2
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr5_+_136058849 | 1.90 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced |
| chr7_-_81770362 | 1.88 |
ENST00000412881.5
|
HGF
|
hepatocyte growth factor |
| chr13_+_75760659 | 1.86 |
ENST00000526202.5
ENST00000465261.6 |
LMO7
|
LIM domain 7 |
| chr1_-_203351115 | 1.84 |
ENST00000354955.5
|
FMOD
|
fibromodulin |
| chr5_+_102865244 | 1.81 |
ENST00000511477.5
ENST00000506006.1 ENST00000509832.5 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_+_202073249 | 1.79 |
ENST00000498697.3
|
KIAA2012
|
KIAA2012 |
| chr4_-_73620391 | 1.77 |
ENST00000395777.6
ENST00000307439.10 |
RASSF6
|
Ras association domain family member 6 |
| chr7_-_81769971 | 1.76 |
ENST00000354224.10
ENST00000643024.1 |
HGF
|
hepatocyte growth factor |
| chr8_+_54559164 | 1.74 |
ENST00000636932.1
|
RP1
|
RP1 axonemal microtubule associated |
| chrX_+_136532205 | 1.74 |
ENST00000370634.8
|
VGLL1
|
vestigial like family member 1 |
| chrX_+_22032301 | 1.74 |
ENST00000379374.5
|
PHEX
|
phosphate regulating endopeptidase homolog X-linked |
| chr12_-_71157872 | 1.63 |
ENST00000546561.2
|
TSPAN8
|
tetraspanin 8 |
| chr2_-_88860913 | 1.63 |
ENST00000390239.2
|
IGKJ4
|
immunoglobulin kappa joining 4 |
| chr1_-_114757958 | 1.62 |
ENST00000525132.1
|
CSDE1
|
cold shock domain containing E1 |
| chr14_+_36657560 | 1.62 |
ENST00000402703.6
|
PAX9
|
paired box 9 |
| chr12_+_103587266 | 1.60 |
ENST00000388887.7
|
STAB2
|
stabilin 2 |
| chr2_+_87748087 | 1.60 |
ENST00000359481.9
|
PLGLB2
|
plasminogen like B2 |
| chr13_+_75760431 | 1.57 |
ENST00000321797.12
|
LMO7
|
LIM domain 7 |
| chr11_+_1157946 | 1.56 |
ENST00000621226.2
|
MUC5AC
|
mucin 5AC, oligomeric mucus/gel-forming |
| chr1_-_116667668 | 1.56 |
ENST00000369486.8
ENST00000369483.5 |
IGSF3
|
immunoglobulin superfamily member 3 |
| chr8_-_132760624 | 1.53 |
ENST00000522334.5
ENST00000519016.5 |
TMEM71
|
transmembrane protein 71 |
| chr11_-_102530738 | 1.53 |
ENST00000260227.5
|
MMP7
|
matrix metallopeptidase 7 |
| chr6_+_10528326 | 1.53 |
ENST00000379597.7
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr19_-_48390847 | 1.53 |
ENST00000597017.5
|
KDELR1
|
KDEL endoplasmic reticulum protein retention receptor 1 |
| chr2_+_70935864 | 1.48 |
ENST00000234396.10
ENST00000454446.6 |
ATP6V1B1
|
ATPase H+ transporting V1 subunit B1 |
| chr15_+_96325935 | 1.42 |
ENST00000421109.6
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chr12_+_120978686 | 1.41 |
ENST00000541395.5
ENST00000544413.2 |
HNF1A
|
HNF1 homeobox A |
| chr16_-_68000549 | 1.41 |
ENST00000575510.5
|
DPEP2
|
dipeptidase 2 |
| chr15_+_71096941 | 1.40 |
ENST00000355327.7
|
THSD4
|
thrombospondin type 1 domain containing 4 |
| chr12_-_102478539 | 1.38 |
ENST00000424202.6
|
IGF1
|
insulin like growth factor 1 |
| chr2_+_70935919 | 1.37 |
ENST00000412314.5
|
ATP6V1B1
|
ATPase H+ transporting V1 subunit B1 |
| chr3_-_146161167 | 1.36 |
ENST00000360060.7
ENST00000282903.10 |
PLOD2
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 |
| chr8_+_94895837 | 1.33 |
ENST00000519136.5
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr21_-_14658812 | 1.32 |
ENST00000647101.1
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr12_-_71157992 | 1.30 |
ENST00000247829.8
|
TSPAN8
|
tetraspanin 8 |
| chr1_-_111200633 | 1.30 |
ENST00000357640.9
|
DENND2D
|
DENN domain containing 2D |
| chr6_-_89315291 | 1.29 |
ENST00000402938.4
|
GABRR2
|
gamma-aminobutyric acid type A receptor subunit rho2 |
| chr6_+_36676489 | 1.27 |
ENST00000448526.6
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr1_-_94925759 | 1.26 |
ENST00000415017.1
ENST00000545882.5 |
CNN3
|
calponin 3 |
| chr17_-_59151794 | 1.25 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr2_+_202073282 | 1.24 |
ENST00000459709.5
|
KIAA2012
|
KIAA2012 |
| chr12_+_120978537 | 1.21 |
ENST00000257555.11
ENST00000400024.6 |
HNF1A
|
HNF1 homeobox A |
| chr1_-_177969907 | 1.20 |
ENST00000308284.10
|
SEC16B
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chrX_+_22032427 | 1.19 |
ENST00000684143.1
|
PHEX
|
phosphate regulating endopeptidase homolog X-linked |
| chr1_+_222928415 | 1.17 |
ENST00000284476.7
|
DISP1
|
dispatched RND transporter family member 1 |
| chr18_+_68798237 | 1.17 |
ENST00000581520.1
|
CCDC102B
|
coiled-coil domain containing 102B |
| chr7_+_114414809 | 1.15 |
ENST00000350908.9
|
FOXP2
|
forkhead box P2 |
| chr4_-_73620629 | 1.15 |
ENST00000342081.7
|
RASSF6
|
Ras association domain family member 6 |
| chr10_-_125161056 | 1.15 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr18_-_25351140 | 1.15 |
ENST00000577461.1
|
ZNF521
|
zinc finger protein 521 |
| chr4_-_110198650 | 1.13 |
ENST00000394607.7
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr7_+_134891566 | 1.12 |
ENST00000424922.5
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr18_-_54959391 | 1.10 |
ENST00000591504.6
|
CCDC68
|
coiled-coil domain containing 68 |
| chr19_-_45406327 | 1.10 |
ENST00000593226.5
ENST00000418234.6 |
PPP1R13L
|
protein phosphatase 1 regulatory subunit 13 like |
| chr14_+_36657664 | 1.09 |
ENST00000555639.2
|
PAX9
|
paired box 9 |
| chr18_+_3449413 | 1.08 |
ENST00000549253.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr14_+_36661852 | 1.08 |
ENST00000361487.7
|
PAX9
|
paired box 9 |
| chr17_-_40984297 | 1.07 |
ENST00000377755.9
|
KRT40
|
keratin 40 |
| chr19_-_48614805 | 1.07 |
ENST00000593772.1
|
FAM83E
|
family with sequence similarity 83 member E |
| chr4_-_73620500 | 1.05 |
ENST00000335049.5
|
RASSF6
|
Ras association domain family member 6 |
| chr7_-_81770428 | 1.05 |
ENST00000421558.1
|
HGF
|
hepatocyte growth factor |
| chr8_+_94895763 | 1.05 |
ENST00000523378.5
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr9_-_20622479 | 1.03 |
ENST00000380338.9
|
MLLT3
|
MLLT3 super elongation complex subunit |
| chr5_+_138873990 | 1.02 |
ENST00000518381.5
|
CTNNA1
|
catenin alpha 1 |
| chr11_+_844406 | 1.02 |
ENST00000397404.5
|
TSPAN4
|
tetraspanin 4 |
| chrX_+_136648138 | 1.02 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand |
| chr5_+_171420016 | 1.00 |
ENST00000675071.1
|
ENSG00000288639.1
|
novel protein |
| chr10_+_102776237 | 1.00 |
ENST00000369889.5
|
WBP1L
|
WW domain binding protein 1 like |
| chr2_+_168901290 | 1.00 |
ENST00000429379.2
ENST00000375363.8 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase catalytic subunit 2 |
| chr2_-_87021844 | 0.98 |
ENST00000355705.4
ENST00000409310.6 |
PLGLB1
|
plasminogen like B1 |
| chr22_-_17219424 | 0.98 |
ENST00000649540.1
ENST00000399837.8 ENST00000543038.1 |
ADA2
|
adenosine deaminase 2 |
| chr4_+_67558719 | 0.96 |
ENST00000265404.7
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr10_+_101131284 | 0.96 |
ENST00000370196.11
ENST00000467928.2 |
TLX1
|
T cell leukemia homeobox 1 |
| chr14_+_22829879 | 0.91 |
ENST00000355151.9
ENST00000397496.7 ENST00000555345.5 ENST00000432849.7 ENST00000553711.5 ENST00000556465.5 ENST00000397505.2 ENST00000557221.1 ENST00000556840.5 ENST00000555536.1 |
MRPL52
|
mitochondrial ribosomal protein L52 |
| chr10_+_113709261 | 0.90 |
ENST00000672138.1
ENST00000452490.3 |
CASP7
|
caspase 7 |
| chr6_-_117425905 | 0.88 |
ENST00000368507.8
|
ROS1
|
ROS proto-oncogene 1, receptor tyrosine kinase |
| chr15_+_59617933 | 0.87 |
ENST00000559200.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr19_+_38389612 | 0.87 |
ENST00000586301.5
|
SPRED3
|
sprouty related EVH1 domain containing 3 |
| chr7_-_93226449 | 0.86 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr3_+_160677152 | 0.86 |
ENST00000320767.4
|
ARL14
|
ADP ribosylation factor like GTPase 14 |
| chr12_+_10212483 | 0.86 |
ENST00000545859.5
|
GABARAPL1
|
GABA type A receptor associated protein like 1 |
| chr3_-_45915596 | 0.85 |
ENST00000472635.5
ENST00000492333.5 |
LZTFL1
|
leucine zipper transcription factor like 1 |
| chr13_+_75760362 | 0.84 |
ENST00000534657.5
|
LMO7
|
LIM domain 7 |
| chr17_+_74935892 | 0.83 |
ENST00000328801.6
|
OTOP3
|
otopetrin 3 |
| chr17_-_41315706 | 0.82 |
ENST00000334202.5
|
KRTAP17-1
|
keratin associated protein 17-1 |
| chr2_-_88860605 | 0.81 |
ENST00000390238.2
|
IGKJ5
|
immunoglobulin kappa joining 5 |
| chr1_+_34759737 | 0.80 |
ENST00000339480.3
|
GJB4
|
gap junction protein beta 4 |
| chr17_-_19748355 | 0.80 |
ENST00000494157.6
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr4_-_40514543 | 0.79 |
ENST00000513473.5
|
RBM47
|
RNA binding motif protein 47 |
| chr3_-_185821092 | 0.79 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr2_+_203936755 | 0.79 |
ENST00000316386.11
ENST00000435193.1 |
ICOS
|
inducible T cell costimulator |
| chr6_+_106098933 | 0.78 |
ENST00000369089.3
|
PRDM1
|
PR/SET domain 1 |
| chr1_-_216423396 | 0.78 |
ENST00000366942.3
ENST00000674083.1 ENST00000307340.8 |
USH2A
|
usherin |
| chr14_+_104801082 | 0.77 |
ENST00000342537.8
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr6_-_107824294 | 0.77 |
ENST00000369020.8
ENST00000369022.6 |
SCML4
|
Scm polycomb group protein like 4 |
| chr21_-_26843012 | 0.77 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chrX_-_20218941 | 0.77 |
ENST00000457145.6
|
RPS6KA3
|
ribosomal protein S6 kinase A3 |
| chr5_+_180494430 | 0.77 |
ENST00000393356.7
ENST00000618123.4 |
CNOT6
|
CCR4-NOT transcription complex subunit 6 |
| chr14_+_104800573 | 0.76 |
ENST00000555360.1
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr12_-_52434363 | 0.76 |
ENST00000252245.6
|
KRT75
|
keratin 75 |
| chr1_-_11047225 | 0.76 |
ENST00000400898.3
ENST00000400897.8 |
MASP2
|
mannan binding lectin serine peptidase 2 |
| chr3_-_114179052 | 0.76 |
ENST00000383673.5
ENST00000295881.9 |
DRD3
|
dopamine receptor D3 |
| chr17_-_17582417 | 0.75 |
ENST00000395783.5
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr4_-_185535498 | 0.75 |
ENST00000284767.12
ENST00000284771.7 ENST00000284770.10 ENST00000620787.5 ENST00000629667.2 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr22_-_17219571 | 0.74 |
ENST00000610390.4
|
ADA2
|
adenosine deaminase 2 |
| chr14_-_64504570 | 0.74 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr4_-_140154176 | 0.74 |
ENST00000509479.6
|
MAML3
|
mastermind like transcriptional coactivator 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.3 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 3.3 | 10.0 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 2.7 | 10.7 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 1.6 | 8.0 | GO:0010193 | response to ozone(GO:0010193) |
| 1.5 | 4.6 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 1.3 | 2.6 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 1.3 | 5.1 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 1.3 | 6.4 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 1.2 | 8.3 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 1.1 | 13.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 1.0 | 3.1 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 1.0 | 13.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.9 | 10.5 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.9 | 2.6 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.8 | 7.6 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.8 | 4.7 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.8 | 3.1 | GO:1900228 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.7 | 14.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.7 | 11.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.7 | 5.2 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.6 | 3.9 | GO:0018032 | protein amidation(GO:0018032) |
| 0.5 | 4.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.5 | 4.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.5 | 1.6 | GO:0021757 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.5 | 2.6 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.5 | 2.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.5 | 2.6 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.5 | 3.0 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.5 | 2.9 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.4 | 2.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.4 | 9.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.4 | 4.9 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.4 | 10.7 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.4 | 1.2 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.4 | 1.8 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.4 | 2.5 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.4 | 1.4 | GO:0009956 | radial pattern formation(GO:0009956) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.3 | 3.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.3 | 2.6 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.3 | 1.6 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.3 | 1.0 | GO:1903970 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.3 | 2.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.3 | 2.8 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 4.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 11.8 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.3 | 1.4 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.3 | 2.4 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.3 | 7.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.3 | 0.8 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.2 | 1.7 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.2 | 0.9 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.2 | 1.3 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.2 | 2.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 2.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 1.5 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 0.8 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.2 | 0.5 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.2 | 6.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 1.0 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 3.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.2 | 0.8 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 4.8 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 6.2 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.1 | 0.6 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 1.7 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 3.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 1.8 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.9 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.5 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 2.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 1.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 8.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 2.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.3 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.7 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 3.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 10.0 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 3.8 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.1 | 1.2 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.1 | 0.8 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 3.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.3 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.5 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 2.9 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 1.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.8 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 1.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 1.5 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.1 | 1.3 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.1 | 1.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.5 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.5 | GO:0051023 | regulation of immunoglobulin secretion(GO:0051023) |
| 0.0 | 0.5 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 1.0 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 3.8 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.7 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 2.9 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.2 | GO:2000174 | pro-T cell differentiation(GO:0002572) regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.6 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.8 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 1.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.8 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 3.3 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.0 | 0.6 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 10.0 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 1.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.5 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.4 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 1.3 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 1.0 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 1.9 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.8 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.3 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.8 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 24.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.4 | 8.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.9 | 7.6 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.8 | 10.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.7 | 22.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.5 | 10.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.5 | 2.9 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.4 | 1.6 | GO:0070701 | mucus layer(GO:0070701) |
| 0.3 | 14.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 8.3 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.3 | 0.8 | GO:0002141 | stereocilia ankle link(GO:0002141) USH2 complex(GO:1990696) |
| 0.3 | 1.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 5.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 3.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 2.9 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 2.4 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 2.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 22.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 2.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 7.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 4.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 3.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.5 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 1.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 8.3 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 9.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 7.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 3.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 1.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 8.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 6.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 7.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 3.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 3.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.7 | GO:1990777 | plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 3.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 31.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 3.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 3.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.6 | 12.7 | GO:0035473 | lipase binding(GO:0035473) |
| 1.3 | 3.8 | GO:0052852 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 1.0 | 3.0 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.9 | 8.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.7 | 7.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.7 | 2.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.6 | 3.2 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.6 | 3.9 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.5 | 8.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.5 | 1.5 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.5 | 1.5 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.4 | 2.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.4 | 4.9 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.4 | 19.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 6.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 1.0 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.3 | 3.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 2.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 0.9 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.3 | 1.4 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.3 | 2.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 1.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.3 | 1.5 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.3 | 0.8 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.2 | 11.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 2.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 0.8 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.2 | 2.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 0.5 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 3.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 3.1 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.2 | 1.7 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.5 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.1 | 1.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 1.8 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 2.8 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 20.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 3.8 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 3.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.8 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 3.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 3.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 8.7 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 0.5 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 4.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 8.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.8 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 5.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 1.2 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 6.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 3.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 2.5 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 1.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.0 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 4.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 17.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 4.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.9 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 28.9 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 1.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 2.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 2.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 5.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.6 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.2 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 5.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 41.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 26.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 17.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 7.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 8.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 6.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 8.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 3.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 24.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 6.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 5.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 3.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 11.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 9.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 2.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.5 | 12.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.5 | 25.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.4 | 13.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 10.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 7.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 3.7 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 6.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.3 | 13.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 7.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 3.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 4.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 2.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 4.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 3.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 6.0 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 2.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 5.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 3.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.0 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 1.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 4.1 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 3.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 1.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.8 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |