Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
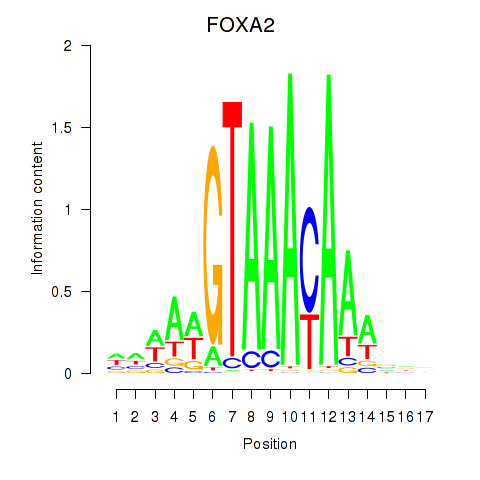
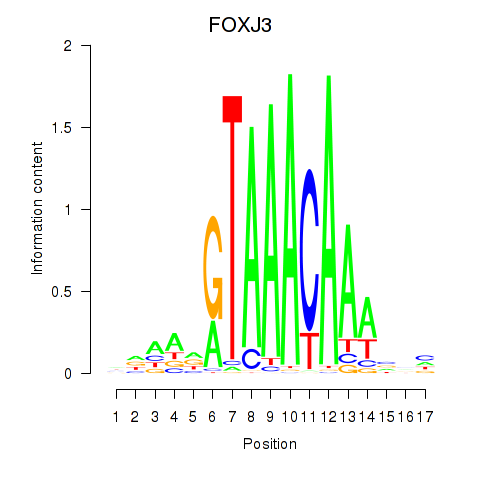
Results for FOXA2_FOXJ3
Z-value: 1.20
Transcription factors associated with FOXA2_FOXJ3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA2
|
ENSG00000125798.15 | forkhead box A2 |
|
FOXJ3
|
ENSG00000198815.9 | forkhead box J3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXJ3 | hg38_v1_chr1_-_42335869_42335891 | -0.56 | 8.6e-04 | Click! |
| FOXA2 | hg38_v1_chr20_-_22585451_22585459 | 0.19 | 3.1e-01 | Click! |
Activity profile of FOXA2_FOXJ3 motif
Sorted Z-values of FOXA2_FOXJ3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_73404255 | 9.63 |
ENST00000621628.4
ENST00000621085.4 ENST00000415165.6 ENST00000295897.9 ENST00000503124.5 ENST00000509063.5 ENST00000401494.7 |
ALB
|
albumin |
| chr1_-_161223559 | 8.30 |
ENST00000469730.2
ENST00000463273.5 ENST00000464492.5 ENST00000367990.7 ENST00000470459.6 ENST00000463812.1 ENST00000468465.5 |
APOA2
|
apolipoprotein A2 |
| chr1_-_161223408 | 5.58 |
ENST00000491350.1
|
APOA2
|
apolipoprotein A2 |
| chr6_+_160702238 | 5.00 |
ENST00000366924.6
ENST00000308192.14 ENST00000418964.1 |
PLG
|
plasminogen |
| chr9_-_101435760 | 4.79 |
ENST00000647789.2
ENST00000616752.1 |
ALDOB
|
aldolase, fructose-bisphosphate B |
| chr1_-_169586539 | 4.45 |
ENST00000367796.3
|
F5
|
coagulation factor V |
| chr1_-_169586471 | 4.30 |
ENST00000367797.9
|
F5
|
coagulation factor V |
| chr1_-_56966133 | 4.28 |
ENST00000535057.5
ENST00000543257.5 |
C8B
|
complement C8 beta chain |
| chr1_-_56966006 | 4.12 |
ENST00000371237.9
|
C8B
|
complement C8 beta chain |
| chr1_-_230745574 | 3.91 |
ENST00000681269.1
|
AGT
|
angiotensinogen |
| chr3_+_109136707 | 3.58 |
ENST00000622536.6
|
C3orf85
|
chromosome 3 open reading frame 85 |
| chrX_+_71215156 | 3.36 |
ENST00000374029.2
ENST00000675209.1 ENST00000647424.1 ENST00000675368.1 ENST00000675609.1 ENST00000646835.1 ENST00000447581.2 |
GJB1
|
gap junction protein beta 1 |
| chr2_+_87748087 | 3.26 |
ENST00000359481.9
|
PLGLB2
|
plasminogen like B2 |
| chr10_+_94938649 | 3.10 |
ENST00000461906.1
ENST00000260682.8 |
CYP2C9
|
cytochrome P450 family 2 subfamily C member 9 |
| chr4_+_186266183 | 3.09 |
ENST00000403665.7
ENST00000492972.6 ENST00000264692.8 |
F11
|
coagulation factor XI |
| chr3_+_98732688 | 3.04 |
ENST00000486334.6
ENST00000394162.5 ENST00000613264.4 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr12_+_20815672 | 3.04 |
ENST00000261196.6
ENST00000381541.7 ENST00000540229.1 |
SLCO1B3
SLCO1B3-SLCO1B7
|
solute carrier organic anion transporter family member 1B3 SLCO1B3-SLCO1B7 readthrough |
| chr1_-_159714581 | 2.73 |
ENST00000255030.9
ENST00000437342.1 ENST00000368112.5 ENST00000368111.5 ENST00000368110.1 |
CRP
|
C-reactive protein |
| chr3_+_98732236 | 2.71 |
ENST00000265261.10
ENST00000483910.5 ENST00000460774.5 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr11_+_118527463 | 2.70 |
ENST00000302783.10
|
TTC36
|
tetratricopeptide repeat domain 36 |
| chr17_-_66220630 | 2.69 |
ENST00000585162.1
|
APOH
|
apolipoprotein H |
| chr4_-_163613505 | 2.63 |
ENST00000339875.9
|
MARCHF1
|
membrane associated ring-CH-type finger 1 |
| chr4_-_69495897 | 2.58 |
ENST00000305107.7
ENST00000639621.1 |
UGT2B4
|
UDP glucuronosyltransferase family 2 member B4 |
| chr10_+_7703300 | 2.40 |
ENST00000358415.9
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr10_+_7703340 | 2.39 |
ENST00000429820.5
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr4_-_69495861 | 2.36 |
ENST00000512583.5
|
UGT2B4
|
UDP glucuronosyltransferase family 2 member B4 |
| chr2_-_87021844 | 2.27 |
ENST00000355705.4
ENST00000409310.6 |
PLGLB1
|
plasminogen like B1 |
| chr14_-_74084393 | 2.25 |
ENST00000350259.8
ENST00000553458.6 |
ALDH6A1
|
aldehyde dehydrogenase 6 family member A1 |
| chr2_+_240868817 | 2.24 |
ENST00000307503.4
|
AGXT
|
alanine--glyoxylate and serine--pyruvate aminotransferase |
| chr10_-_100065394 | 2.12 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N subunit 1 |
| chr6_-_160664270 | 1.92 |
ENST00000316300.10
|
LPA
|
lipoprotein(a) |
| chr8_-_80080816 | 1.87 |
ENST00000520527.5
ENST00000517427.5 ENST00000379097.7 ENST00000448733.3 |
TPD52
|
tumor protein D52 |
| chr3_+_98732430 | 1.87 |
ENST00000497008.5
|
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr4_-_139302516 | 1.77 |
ENST00000394228.5
ENST00000539387.5 |
NDUFC1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr2_+_165239432 | 1.73 |
ENST00000636071.2
ENST00000636985.2 |
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr8_+_119067239 | 1.70 |
ENST00000332843.3
|
COLEC10
|
collectin subfamily member 10 |
| chr16_+_56191536 | 1.67 |
ENST00000569295.6
|
GNAO1
|
G protein subunit alpha o1 |
| chr16_+_6019585 | 1.66 |
ENST00000547372.5
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr12_-_95996302 | 1.64 |
ENST00000261208.8
ENST00000538703.5 ENST00000541929.5 |
HAL
|
histidine ammonia-lyase |
| chr1_+_47137435 | 1.62 |
ENST00000371891.8
ENST00000371890.7 ENST00000619754.4 ENST00000294337.7 ENST00000620131.1 |
CYP4A22
|
cytochrome P450 family 4 subfamily A member 22 |
| chr2_-_71227055 | 1.56 |
ENST00000244221.9
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr17_+_7583828 | 1.55 |
ENST00000396501.8
ENST00000250124.11 ENST00000584378.5 ENST00000423172.6 ENST00000579445.5 ENST00000585217.5 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr14_+_88005128 | 1.55 |
ENST00000267549.5
|
GPR65
|
G protein-coupled receptor 65 |
| chr1_-_157841800 | 1.54 |
ENST00000368174.5
|
CD5L
|
CD5 molecule like |
| chr4_+_89111521 | 1.51 |
ENST00000603357.3
|
TIGD2
|
tigger transposable element derived 2 |
| chr5_-_41213021 | 1.50 |
ENST00000417809.1
|
C6
|
complement C6 |
| chr2_+_142877653 | 1.49 |
ENST00000375773.6
ENST00000409512.5 ENST00000264170.9 ENST00000410015.6 |
KYNU
|
kynureninase |
| chr2_+_165239388 | 1.49 |
ENST00000424833.5
ENST00000375437.7 ENST00000631182.3 |
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chrX_+_49162564 | 1.44 |
ENST00000616266.4
|
MAGIX
|
MAGI family member, X-linked |
| chr6_+_122789197 | 1.40 |
ENST00000368440.5
|
SMPDL3A
|
sphingomyelin phosphodiesterase acid like 3A |
| chr9_+_72114595 | 1.40 |
ENST00000545168.5
|
GDA
|
guanine deaminase |
| chr16_+_4616475 | 1.38 |
ENST00000591895.5
|
MGRN1
|
mahogunin ring finger 1 |
| chr16_+_6019016 | 1.37 |
ENST00000550418.6
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr1_+_181483541 | 1.37 |
ENST00000367570.5
ENST00000621791.4 |
CACNA1E
|
calcium voltage-gated channel subunit alpha1 E |
| chr4_-_139302460 | 1.32 |
ENST00000394223.2
ENST00000676245.1 |
NDUFC1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr15_-_53759634 | 1.32 |
ENST00000557913.5
ENST00000360509.10 |
WDR72
|
WD repeat domain 72 |
| chrX_+_10156960 | 1.31 |
ENST00000380833.9
|
CLCN4
|
chloride voltage-gated channel 4 |
| chr1_-_246566238 | 1.30 |
ENST00000366514.5
|
TFB2M
|
transcription factor B2, mitochondrial |
| chr4_+_118034480 | 1.28 |
ENST00000296499.6
|
NDST3
|
N-deacetylase and N-sulfotransferase 3 |
| chr1_-_206921867 | 1.28 |
ENST00000628511.2
ENST00000367091.8 |
FCMR
|
Fc fragment of IgM receptor |
| chr16_+_6019071 | 1.26 |
ENST00000547605.5
ENST00000553186.5 |
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr11_+_126327863 | 1.24 |
ENST00000648516.1
|
DCPS
|
decapping enzyme, scavenger |
| chr10_-_127892930 | 1.24 |
ENST00000368671.4
|
CLRN3
|
clarin 3 |
| chr7_+_26293025 | 1.22 |
ENST00000396376.5
|
SNX10
|
sorting nexin 10 |
| chr8_+_66432475 | 1.21 |
ENST00000415254.5
ENST00000396623.8 |
ADHFE1
|
alcohol dehydrogenase iron containing 1 |
| chr2_+_113127588 | 1.20 |
ENST00000409930.4
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr13_-_99258366 | 1.20 |
ENST00000397470.5
ENST00000397473.7 |
GPR18
|
G protein-coupled receptor 18 |
| chr2_-_2326210 | 1.19 |
ENST00000647755.1
|
MYT1L
|
myelin transcription factor 1 like |
| chr1_+_50105666 | 1.19 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr3_+_186974957 | 1.17 |
ENST00000438590.5
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr16_-_88706262 | 1.16 |
ENST00000562544.1
|
RNF166
|
ring finger protein 166 |
| chr1_+_181483510 | 1.15 |
ENST00000367573.7
|
CACNA1E
|
calcium voltage-gated channel subunit alpha1 E |
| chr2_-_178702479 | 1.15 |
ENST00000414766.5
|
TTN
|
titin |
| chr1_+_207496229 | 1.14 |
ENST00000367051.6
ENST00000367053.6 ENST00000367052.6 |
CR1
|
complement C3b/C4b receptor 1 (Knops blood group) |
| chr4_-_110198976 | 1.10 |
ENST00000506625.5
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr16_+_56191476 | 1.09 |
ENST00000262493.12
|
GNAO1
|
G protein subunit alpha o1 |
| chr7_+_99828010 | 1.09 |
ENST00000631161.2
ENST00000354829.7 ENST00000342499.8 ENST00000417625.5 ENST00000415413.5 ENST00000444905.5 ENST00000222382.5 ENST00000312017.9 |
CYP3A43
|
cytochrome P450 family 3 subfamily A member 43 |
| chr4_+_87799546 | 1.09 |
ENST00000226284.7
|
IBSP
|
integrin binding sialoprotein |
| chr1_-_206921987 | 1.08 |
ENST00000530505.1
ENST00000442471.4 |
FCMR
|
Fc fragment of IgM receptor |
| chr19_-_51751854 | 1.08 |
ENST00000304748.5
ENST00000595042.5 |
FPR1
|
formyl peptide receptor 1 |
| chr2_+_174395721 | 1.06 |
ENST00000272732.11
ENST00000458563.5 ENST00000409673.7 ENST00000435964.1 ENST00000424069.5 ENST00000427038.5 |
SCRN3
|
secernin 3 |
| chr12_-_12561091 | 1.02 |
ENST00000539940.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr1_+_207496268 | 1.02 |
ENST00000529814.1
|
CR1
|
complement C3b/C4b receptor 1 (Knops blood group) |
| chr1_-_160523204 | 1.02 |
ENST00000368055.1
ENST00000368057.8 ENST00000368059.7 |
SLAMF6
|
SLAM family member 6 |
| chr12_-_95995920 | 1.01 |
ENST00000552509.5
|
HAL
|
histidine ammonia-lyase |
| chr5_-_36241798 | 1.01 |
ENST00000514504.5
|
NADK2
|
NAD kinase 2, mitochondrial |
| chr1_-_216805367 | 1.00 |
ENST00000360012.7
|
ESRRG
|
estrogen related receptor gamma |
| chr12_-_12684490 | 0.99 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr4_-_175891691 | 0.99 |
ENST00000507540.1
|
GPM6A
|
glycoprotein M6A |
| chr17_-_41309253 | 0.96 |
ENST00000391352.1
|
KRTAP16-1
|
keratin associated protein 16-1 |
| chr14_+_104801082 | 0.96 |
ENST00000342537.8
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr4_+_87832917 | 0.95 |
ENST00000395102.8
ENST00000497649.6 ENST00000540395.1 ENST00000560249.5 ENST00000511670.5 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr5_+_76875177 | 0.95 |
ENST00000613039.1
|
S100Z
|
S100 calcium binding protein Z |
| chr3_-_58537283 | 0.95 |
ENST00000459701.6
|
ACOX2
|
acyl-CoA oxidase 2 |
| chr1_+_241532370 | 0.95 |
ENST00000366559.9
ENST00000366557.8 |
KMO
|
kynurenine 3-monooxygenase |
| chr10_-_50279715 | 0.94 |
ENST00000395526.9
|
ASAH2
|
N-acylsphingosine amidohydrolase 2 |
| chr5_-_36241875 | 0.93 |
ENST00000381937.9
|
NADK2
|
NAD kinase 2, mitochondrial |
| chr4_-_110199173 | 0.92 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr15_+_58138368 | 0.92 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chr1_+_44405164 | 0.91 |
ENST00000355387.6
|
RNF220
|
ring finger protein 220 |
| chr12_-_12561072 | 0.91 |
ENST00000541207.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr2_-_2326378 | 0.89 |
ENST00000647618.1
|
MYT1L
|
myelin transcription factor 1 like |
| chr18_+_49562049 | 0.89 |
ENST00000261292.9
ENST00000427224.6 ENST00000580036.5 |
LIPG
|
lipase G, endothelial type |
| chr12_+_28452493 | 0.88 |
ENST00000542801.5
|
CCDC91
|
coiled-coil domain containing 91 |
| chr1_+_207496147 | 0.88 |
ENST00000400960.7
ENST00000367049.9 |
CR1
|
complement C3b/C4b receptor 1 (Knops blood group) |
| chr8_-_40897814 | 0.88 |
ENST00000297737.11
ENST00000315769.11 |
ZMAT4
|
zinc finger matrin-type 4 |
| chr1_+_116754422 | 0.86 |
ENST00000369478.4
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr15_-_40307910 | 0.86 |
ENST00000543785.3
ENST00000260402.8 |
PLCB2
|
phospholipase C beta 2 |
| chr12_-_102197827 | 0.86 |
ENST00000329406.5
|
PMCH
|
pro-melanin concentrating hormone |
| chr5_-_36241944 | 0.85 |
ENST00000511088.1
ENST00000282512.7 ENST00000506945.5 |
NADK2
|
NAD kinase 2, mitochondrial |
| chr20_-_1994046 | 0.85 |
ENST00000217305.3
ENST00000650874.1 |
PDYN
|
prodynorphin |
| chr15_-_65029576 | 0.82 |
ENST00000220058.9
|
MTFMT
|
mitochondrial methionyl-tRNA formyltransferase |
| chr3_+_190615308 | 0.82 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr3_-_58537181 | 0.82 |
ENST00000302819.10
|
ACOX2
|
acyl-CoA oxidase 2 |
| chr15_-_40307825 | 0.82 |
ENST00000456256.6
ENST00000557821.5 |
PLCB2
|
phospholipase C beta 2 |
| chr8_+_86342539 | 0.80 |
ENST00000517970.6
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr6_+_101398788 | 0.80 |
ENST00000369138.5
ENST00000413795.5 ENST00000358361.7 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr2_-_206213430 | 0.80 |
ENST00000447845.1
|
GPR1
|
G protein-coupled receptor 1 |
| chrX_+_121047601 | 0.80 |
ENST00000328078.3
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr6_+_10528326 | 0.79 |
ENST00000379597.7
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr15_-_81324130 | 0.79 |
ENST00000302824.7
|
STARD5
|
StAR related lipid transfer domain containing 5 |
| chr3_+_155083889 | 0.78 |
ENST00000680282.1
|
MME
|
membrane metalloendopeptidase |
| chr1_+_15526813 | 0.77 |
ENST00000375838.5
ENST00000616884.4 ENST00000375849.5 ENST00000375847.8 |
DNAJC16
|
DnaJ heat shock protein family (Hsp40) member C16 |
| chr15_+_58138169 | 0.76 |
ENST00000558772.5
|
AQP9
|
aquaporin 9 |
| chr1_+_198638457 | 0.76 |
ENST00000367379.6
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr3_+_155024118 | 0.75 |
ENST00000492661.5
|
MME
|
membrane metalloendopeptidase |
| chr6_-_25830557 | 0.74 |
ENST00000468082.1
|
SLC17A1
|
solute carrier family 17 member 1 |
| chr3_-_129656713 | 0.74 |
ENST00000510323.1
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr12_-_51324652 | 0.74 |
ENST00000544402.5
|
BIN2
|
bridging integrator 2 |
| chr14_+_104800573 | 0.74 |
ENST00000555360.1
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr17_+_7627963 | 0.73 |
ENST00000575729.5
ENST00000340624.9 |
SHBG
|
sex hormone binding globulin |
| chr11_-_126268810 | 0.73 |
ENST00000332118.11
|
SRPRA
|
SRP receptor subunit alpha |
| chr18_+_23453275 | 0.72 |
ENST00000581585.5
ENST00000339486.8 ENST00000577501.5 |
RIOK3
|
RIO kinase 3 |
| chr8_-_63026179 | 0.71 |
ENST00000677919.1
|
GGH
|
gamma-glutamyl hydrolase |
| chr4_-_64409444 | 0.70 |
ENST00000381210.8
ENST00000507440.5 |
TECRL
|
trans-2,3-enoyl-CoA reductase like |
| chr16_+_48244331 | 0.70 |
ENST00000535754.5
|
LONP2
|
lon peptidase 2, peroxisomal |
| chr19_+_54906140 | 0.70 |
ENST00000291890.9
ENST00000598576.5 ENST00000594765.5 ENST00000350790.9 ENST00000338835.9 ENST00000357397.5 |
NCR1
|
natural cytotoxicity triggering receptor 1 |
| chr16_+_48244264 | 0.70 |
ENST00000285737.9
|
LONP2
|
lon peptidase 2, peroxisomal |
| chr2_-_206213362 | 0.70 |
ENST00000442134.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr4_+_67558719 | 0.70 |
ENST00000265404.7
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr4_-_64409381 | 0.69 |
ENST00000509536.1
|
TECRL
|
trans-2,3-enoyl-CoA reductase like |
| chr14_-_60165293 | 0.69 |
ENST00000554101.5
ENST00000557137.1 |
DHRS7
|
dehydrogenase/reductase 7 |
| chr4_+_116298876 | 0.69 |
ENST00000604093.2
|
MTRNR2L13
|
MT-RNR2 like 13 |
| chr1_+_174877430 | 0.69 |
ENST00000392064.6
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr10_-_67838019 | 0.69 |
ENST00000483798.6
|
DNAJC12
|
DnaJ heat shock protein family (Hsp40) member C12 |
| chr3_+_119597874 | 0.68 |
ENST00000488919.5
ENST00000273371.9 ENST00000495992.5 |
PLA1A
|
phospholipase A1 member A |
| chr16_+_7332744 | 0.68 |
ENST00000436368.6
ENST00000311745.9 ENST00000340209.8 ENST00000620507.4 |
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr5_+_65926644 | 0.68 |
ENST00000511297.5
ENST00000506030.5 |
ERBIN
|
erbb2 interacting protein |
| chr18_-_50825373 | 0.68 |
ENST00000588444.5
ENST00000256425.6 ENST00000428869.6 |
MRO
|
maestro |
| chr12_-_54981838 | 0.68 |
ENST00000316577.12
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr19_-_50637939 | 0.67 |
ENST00000338916.8
|
SYT3
|
synaptotagmin 3 |
| chr18_+_57352541 | 0.67 |
ENST00000324000.4
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr4_-_110198650 | 0.67 |
ENST00000394607.7
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr15_+_90191913 | 0.66 |
ENST00000559792.5
|
SEMA4B
|
semaphorin 4B |
| chr16_+_10386049 | 0.66 |
ENST00000562527.5
ENST00000396559.5 ENST00000396560.6 ENST00000562102.5 ENST00000543967.5 ENST00000569939.5 ENST00000569900.5 |
ATF7IP2
|
activating transcription factor 7 interacting protein 2 |
| chr5_+_167754918 | 0.66 |
ENST00000519204.5
|
TENM2
|
teneurin transmembrane protein 2 |
| chr4_-_40630826 | 0.65 |
ENST00000505414.5
ENST00000511598.5 |
RBM47
|
RNA binding motif protein 47 |
| chr6_+_146029059 | 0.65 |
ENST00000282753.6
|
GRM1
|
glutamate metabotropic receptor 1 |
| chr2_-_36966503 | 0.64 |
ENST00000263918.9
|
STRN
|
striatin |
| chr19_-_51804104 | 0.64 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr1_+_198638723 | 0.64 |
ENST00000643513.1
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr4_-_69214743 | 0.63 |
ENST00000446444.2
|
UGT2B11
|
UDP glucuronosyltransferase family 2 member B11 |
| chr16_-_28623560 | 0.63 |
ENST00000350842.8
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chrX_-_77895546 | 0.63 |
ENST00000358075.11
|
MAGT1
|
magnesium transporter 1 |
| chrY_-_23694579 | 0.62 |
ENST00000343584.10
|
PRYP3
|
PTPN13 like Y-linked pseudogene 3 |
| chr6_-_127900958 | 0.61 |
ENST00000434358.3
ENST00000630369.2 ENST00000368248.4 |
THEMIS
|
thymocyte selection associated |
| chr19_+_56176006 | 0.60 |
ENST00000357330.7
ENST00000440823.1 |
GALP
|
galanin like peptide |
| chr17_-_3691887 | 0.60 |
ENST00000552050.5
|
P2RX5
|
purinergic receptor P2X 5 |
| chr6_+_96924720 | 0.60 |
ENST00000369261.9
|
KLHL32
|
kelch like family member 32 |
| chr14_-_100569780 | 0.60 |
ENST00000355173.7
|
BEGAIN
|
brain enriched guanylate kinase associated |
| chr2_-_169573766 | 0.60 |
ENST00000438035.5
ENST00000453929.6 |
FASTKD1
|
FAST kinase domains 1 |
| chr7_+_26291941 | 0.60 |
ENST00000412416.5
|
SNX10
|
sorting nexin 10 |
| chr10_-_97334698 | 0.60 |
ENST00000371019.4
|
FRAT2
|
FRAT regulator of WNT signaling pathway 2 |
| chr10_-_67838173 | 0.60 |
ENST00000225171.7
|
DNAJC12
|
DnaJ heat shock protein family (Hsp40) member C12 |
| chr19_+_3880647 | 0.60 |
ENST00000450849.7
|
ATCAY
|
ATCAY kinesin light chain interacting caytaxin |
| chr1_+_198639162 | 0.59 |
ENST00000418674.1
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr10_+_122980448 | 0.59 |
ENST00000405485.2
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr12_+_53985065 | 0.59 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr4_+_109848102 | 0.58 |
ENST00000594814.6
|
LRIT3
|
leucine rich repeat, Ig-like and transmembrane domains 3 |
| chr10_-_124093582 | 0.58 |
ENST00000462406.1
ENST00000435907.6 |
CHST15
|
carbohydrate sulfotransferase 15 |
| chr21_-_15002364 | 0.58 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr1_-_67054100 | 0.58 |
ENST00000235345.6
|
SLC35D1
|
solute carrier family 35 member D1 |
| chr5_+_161848314 | 0.58 |
ENST00000437025.6
|
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr19_-_14674886 | 0.57 |
ENST00000344373.8
ENST00000595472.1 |
ADGRE3
|
adhesion G protein-coupled receptor E3 |
| chr1_+_74235377 | 0.57 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr5_+_167754997 | 0.57 |
ENST00000520394.5
|
TENM2
|
teneurin transmembrane protein 2 |
| chrX_+_78747705 | 0.57 |
ENST00000614823.5
ENST00000435339.3 ENST00000514744.5 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr3_-_151203201 | 0.56 |
ENST00000480322.1
ENST00000309180.6 |
GPR171
|
G protein-coupled receptor 171 |
| chr19_+_18097763 | 0.56 |
ENST00000262811.10
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr15_-_58749569 | 0.56 |
ENST00000402627.5
ENST00000559053.1 ENST00000260408.8 ENST00000561288.1 ENST00000461408.2 ENST00000439637.5 ENST00000558004.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr12_+_71686473 | 0.55 |
ENST00000549735.5
|
TMEM19
|
transmembrane protein 19 |
| chr3_+_155083523 | 0.55 |
ENST00000680057.1
|
MME
|
membrane metalloendopeptidase |
| chr9_+_124291935 | 0.55 |
ENST00000546191.5
|
NEK6
|
NIMA related kinase 6 |
| chr13_-_61415508 | 0.54 |
ENST00000409204.4
|
PCDH20
|
protocadherin 20 |
| chr4_+_183905266 | 0.54 |
ENST00000308497.9
|
STOX2
|
storkhead box 2 |
| chr8_-_144529048 | 0.54 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.6 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chrX_+_86714623 | 0.54 |
ENST00000484479.1
|
DACH2
|
dachshund family transcription factor 2 |
| chr1_+_44674688 | 0.53 |
ENST00000418644.5
ENST00000458657.6 ENST00000535358.6 ENST00000441519.5 ENST00000445071.5 |
ARMH1
|
armadillo like helical domain containing 1 |
| chr12_-_62191422 | 0.53 |
ENST00000551449.1
|
TAFA2
|
TAFA chemokine like family member 2 |
| chr2_-_227614840 | 0.53 |
ENST00000642029.1
|
SCYGR3
|
small cysteine and glycine repeat containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA2_FOXJ3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 13.9 | GO:2000910 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 3.2 | 9.6 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 2.0 | 8.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 1.6 | 4.9 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 1.5 | 8.8 | GO:0032571 | response to vitamin K(GO:0032571) |
| 1.3 | 3.9 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 1.0 | 3.0 | GO:0045957 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.6 | 2.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.6 | 2.2 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.5 | 1.5 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.5 | 1.5 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.5 | 1.9 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.5 | 4.8 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.5 | 1.4 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.5 | 2.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.5 | 7.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 3.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.4 | 1.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.4 | 2.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.4 | 1.2 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.4 | 1.2 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.4 | 1.2 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.4 | 2.6 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.4 | 1.1 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 0.3 | 2.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.3 | 1.0 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.3 | 1.7 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.3 | 1.6 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.2 | 0.7 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.2 | 0.7 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.2 | 0.9 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 0.7 | GO:1903970 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.2 | 0.7 | GO:0045175 | basal protein localization(GO:0045175) |
| 0.2 | 2.7 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) negative regulation of fibrinolysis(GO:0051918) |
| 0.2 | 0.9 | GO:0010983 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.2 | 1.0 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.2 | 0.6 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.2 | 2.7 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 3.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 | 0.6 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.2 | 0.5 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.2 | 0.5 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.2 | 1.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.2 | 8.6 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 2.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.2 | 1.5 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.2 | 0.5 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.2 | 0.6 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.1 | 1.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.6 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 | 0.4 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 0.7 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 3.4 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 2.7 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.1 | 1.7 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 1.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.4 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.4 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 2.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 1.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.5 | GO:1903217 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.1 | 0.7 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.6 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 0.3 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.8 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.5 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.1 | 0.4 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.1 | 0.3 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.1 | 0.3 | GO:0042309 | homoiothermy(GO:0042309) |
| 0.1 | 0.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 2.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.3 | GO:0035928 | mitochondrial RNA 5'-end processing(GO:0000964) rRNA import into mitochondrion(GO:0035928) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.1 | 1.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 2.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.9 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.1 | 1.7 | GO:0032740 | positive regulation of interleukin-17 production(GO:0032740) |
| 0.1 | 0.7 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.1 | 0.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.2 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.1 | 0.3 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.3 | GO:0070510 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.1 | 0.3 | GO:0045014 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 1.9 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.2 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.1 | 0.5 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 3.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.2 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.1 | 0.4 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 5.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.4 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.3 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.1 | 0.8 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.1 | 3.1 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.1 | 0.6 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) UDP-glucuronate metabolic process(GO:0046398) |
| 0.1 | 1.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.3 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.1 | 1.5 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.9 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 0.3 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 1.5 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.3 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.1 | 0.5 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.5 | GO:1902261 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 4.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:1903758 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.2 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 5.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.4 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.5 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.7 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 2.8 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.2 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 3.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 5.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 2.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.3 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 1.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 1.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.5 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:0036404 | positive regulation of Schwann cell differentiation(GO:0014040) conversion of ds siRNA to ss siRNA involved in RNA interference(GO:0033168) conversion of ds siRNA to ss siRNA(GO:0036404) |
| 0.0 | 0.7 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.0 | 0.3 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.5 | GO:0002858 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.0 | 0.6 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.4 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.2 | GO:0035603 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.0 | 0.2 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 1.2 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.2 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 1.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 1.1 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.8 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.4 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.3 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.4 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.6 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.4 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.4 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.7 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.2 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:0060903 | regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 1.0 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.7 | GO:0007035 | vacuolar acidification(GO:0007035) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.7 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.7 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.5 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 0.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.6 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.1 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.9 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.8 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.0 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.1 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.7 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 4.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.0 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.0 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0046851 | negative regulation of bone resorption(GO:0045779) negative regulation of bone remodeling(GO:0046851) |
| 0.0 | 0.4 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 9.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.9 | 5.3 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.7 | 13.9 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.4 | 1.5 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.2 | 1.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 18.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.6 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 2.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 3.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 3.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 4.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.5 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 6.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.3 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.5 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.1 | 0.9 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 2.0 | GO:1990777 | plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.1 | 0.3 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 1.0 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 3.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 5.6 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0033167 | ARC complex(GO:0033167) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 2.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 1.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 1.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 5.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.7 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.0 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 2.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.0 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 2.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 6.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.4 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 12.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 9.5 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 13.9 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.8 | 5.5 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 1.5 | 7.6 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 1.2 | 4.8 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 1.0 | 3.0 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.7 | 2.2 | GO:0033791 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholestanoyl-CoA 24-hydroxylase activity(GO:0033791) |
| 0.7 | 2.7 | GO:0033265 | choline binding(GO:0033265) |
| 0.6 | 3.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.5 | 1.4 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.4 | 2.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.4 | 9.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.4 | 1.2 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.4 | 2.6 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.4 | 2.2 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.3 | 1.0 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.3 | 1.7 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.3 | 1.5 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 1.7 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.2 | 0.7 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.2 | 0.6 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.2 | 0.8 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.2 | 2.7 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 1.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 1.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 2.8 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 0.5 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.2 | 2.6 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) |
| 0.2 | 3.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 3.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 1.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 5.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 1.3 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 0.4 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.1 | 0.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 3.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.4 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 1.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.5 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 1.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.5 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 0.5 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 5.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.7 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.3 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.1 | 0.2 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.1 | 0.6 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.8 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 1.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.3 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 1.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.9 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.7 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 1.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.7 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.6 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 0.7 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 0.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 8.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 3.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.7 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.0 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 1.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.9 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.9 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.7 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 0.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.6 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 1.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 1.9 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 1.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 3.0 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 2.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 2.0 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 0.6 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 3.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.6 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.3 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.1 | 0.3 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 0.5 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 1.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.8 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 2.1 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 2.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.6 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 1.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.7 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.8 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.6 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.8 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 1.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 1.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 1.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.4 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 1.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.7 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 1.0 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 4.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 1.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.6 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.7 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.4 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 1.4 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 1.0 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.0 | GO:0008940 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 3.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 14.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 6.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 2.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 2.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 2.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.6 | 13.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.5 | 8.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 16.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 8.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 6.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 5.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 2.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 3.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 3.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.9 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 2.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 3.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 0.8 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 0.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 4.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 4.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.0 | 1.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 2.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 5.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.5 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.8 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 0.6 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 2.4 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 4.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.4 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 1.1 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 3.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.4 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |