Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
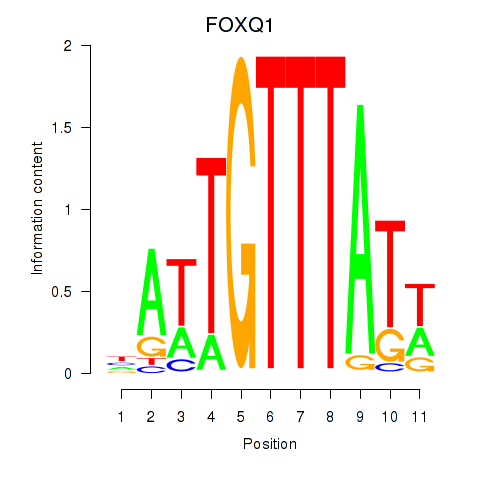
Results for FOXQ1
Z-value: 1.21
Transcription factors associated with FOXQ1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXQ1
|
ENSG00000164379.7 | forkhead box Q1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXQ1 | hg38_v1_chr6_+_1312090_1312105 | 0.06 | 7.5e-01 | Click! |
Activity profile of FOXQ1 motif
Sorted Z-values of FOXQ1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_52803807 | 3.74 |
ENST00000334575.6
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr6_-_52909666 | 2.85 |
ENST00000370968.5
ENST00000211122.4 |
GSTA3
|
glutathione S-transferase alpha 3 |
| chr20_+_33217325 | 2.66 |
ENST00000375452.3
ENST00000375454.8 |
BPIFA3
|
BPI fold containing family A member 3 |
| chr6_-_52763473 | 2.58 |
ENST00000493422.3
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr4_+_154563003 | 2.42 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chrX_+_105948429 | 2.17 |
ENST00000540278.1
|
NRK
|
Nik related kinase |
| chr6_+_131637296 | 2.15 |
ENST00000358229.6
ENST00000357639.8 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr7_-_41703062 | 2.14 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chrX_+_107777733 | 2.01 |
ENST00000509000.3
|
NCBP2L
|
nuclear cap binding protein subunit 2 like |
| chr14_+_21030201 | 1.98 |
ENST00000321760.11
ENST00000460647.6 ENST00000530140.6 ENST00000472458.5 |
TPPP2
|
tubulin polymerization promoting protein family member 2 |
| chr6_-_87095059 | 1.95 |
ENST00000369582.6
ENST00000610310.3 ENST00000630630.2 ENST00000627148.3 ENST00000625577.1 |
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr6_-_49744378 | 1.92 |
ENST00000371159.8
ENST00000263045.9 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr3_+_107377433 | 1.89 |
ENST00000261058.3
|
CCDC54
|
coiled-coil domain containing 54 |
| chrX_-_55182442 | 1.83 |
ENST00000545075.3
|
MTRNR2L10
|
MT-RNR2 like 10 |
| chrX_+_111876177 | 1.73 |
ENST00000635763.2
|
TRPC5OS
|
TRPC5 opposite strand |
| chr6_-_49744434 | 1.72 |
ENST00000433368.6
ENST00000354620.4 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr1_+_196819731 | 1.65 |
ENST00000320493.10
ENST00000367424.4 |
CFHR1
|
complement factor H related 1 |
| chr1_-_237945275 | 1.52 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2 like 11 |
| chr9_-_114074969 | 1.51 |
ENST00000466610.6
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr3_+_171843337 | 1.49 |
ENST00000334567.9
ENST00000619900.4 ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr11_+_73950985 | 1.49 |
ENST00000339764.6
|
DNAJB13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr11_+_7088991 | 1.46 |
ENST00000306904.7
|
RBMXL2
|
RBMX like 2 |
| chr9_-_95317671 | 1.45 |
ENST00000490972.7
ENST00000647778.1 ENST00000649611.1 ENST00000289081.8 |
FANCC
|
FA complementation group C |
| chrX_+_9912434 | 1.45 |
ENST00000418909.6
|
SHROOM2
|
shroom family member 2 |
| chr11_-_26572102 | 1.41 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated |
| chr7_-_122702912 | 1.41 |
ENST00000447240.1
ENST00000434824.2 |
RNF148
|
ring finger protein 148 |
| chr8_-_48921419 | 1.39 |
ENST00000020945.4
|
SNAI2
|
snail family transcriptional repressor 2 |
| chrX_+_108044967 | 1.38 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr17_+_9576627 | 1.36 |
ENST00000396219.7
ENST00000352665.10 |
CFAP52
|
cilia and flagella associated protein 52 |
| chr7_+_128709052 | 1.36 |
ENST00000485070.5
|
FAM71F1
|
family with sequence similarity 71 member F1 |
| chrX_+_111876051 | 1.34 |
ENST00000612026.5
|
TRPC5OS
|
TRPC5 opposite strand |
| chr4_-_118352967 | 1.32 |
ENST00000296498.3
|
PRSS12
|
serine protease 12 |
| chrX_+_108045050 | 1.31 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr7_-_122699108 | 1.31 |
ENST00000340112.3
|
RNF133
|
ring finger protein 133 |
| chr15_+_50182188 | 1.26 |
ENST00000267842.10
|
SLC27A2
|
solute carrier family 27 member 2 |
| chr10_+_125973373 | 1.26 |
ENST00000417114.5
ENST00000445510.5 ENST00000368691.5 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr3_-_57199938 | 1.26 |
ENST00000473921.2
ENST00000295934.8 |
HESX1
|
HESX homeobox 1 |
| chr1_+_197912462 | 1.25 |
ENST00000475727.1
ENST00000367391.5 ENST00000367390.7 |
LHX9
|
LIM homeobox 9 |
| chr1_+_196943738 | 1.25 |
ENST00000367415.8
ENST00000367421.5 ENST00000649283.1 ENST00000476712.6 ENST00000496448.6 ENST00000473386.1 ENST00000649960.1 |
CFHR2
|
complement factor H related 2 |
| chr15_+_50182215 | 1.23 |
ENST00000380902.8
|
SLC27A2
|
solute carrier family 27 member 2 |
| chr12_-_10884244 | 1.20 |
ENST00000543626.4
|
PRH1
|
proline rich protein HaeIII subfamily 1 |
| chr11_-_26572254 | 1.19 |
ENST00000529533.6
|
MUC15
|
mucin 15, cell surface associated |
| chr15_+_75195613 | 1.18 |
ENST00000563905.5
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr3_+_88338451 | 1.18 |
ENST00000637986.2
|
CSNKA2IP
|
casein kinase 2 subunit alpha' interacting protein |
| chr2_+_79025678 | 1.18 |
ENST00000393897.6
|
REG3G
|
regenerating family member 3 gamma |
| chr2_+_79025696 | 1.16 |
ENST00000272324.10
|
REG3G
|
regenerating family member 3 gamma |
| chr12_-_91180365 | 1.16 |
ENST00000547937.5
|
DCN
|
decorin |
| chr1_+_151766655 | 1.16 |
ENST00000400999.7
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr11_-_26572130 | 1.15 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr1_-_75932392 | 1.14 |
ENST00000284142.7
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr3_-_173141227 | 1.14 |
ENST00000351008.4
|
SPATA16
|
spermatogenesis associated 16 |
| chr9_+_69820827 | 1.13 |
ENST00000527647.5
ENST00000480564.1 |
C9orf135
|
chromosome 9 open reading frame 135 |
| chr4_+_122378966 | 1.12 |
ENST00000446706.5
ENST00000296513.7 |
ADAD1
|
adenosine deaminase domain containing 1 |
| chr6_+_31137646 | 1.10 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr10_-_96271553 | 1.07 |
ENST00000224337.10
|
BLNK
|
B cell linker |
| chr16_+_4734519 | 1.07 |
ENST00000299320.10
|
C16orf71
|
chromosome 16 open reading frame 71 |
| chr10_-_11532275 | 1.07 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like |
| chr12_-_91111460 | 1.05 |
ENST00000266718.5
|
LUM
|
lumican |
| chr14_+_77377115 | 1.05 |
ENST00000216471.4
|
SAMD15
|
sterile alpha motif domain containing 15 |
| chr16_+_4734457 | 1.05 |
ENST00000590191.1
|
C16orf71
|
chromosome 16 open reading frame 71 |
| chr19_+_49930219 | 1.04 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr6_-_49866527 | 1.03 |
ENST00000335847.9
|
CRISP1
|
cysteine rich secretory protein 1 |
| chr20_+_13008919 | 1.03 |
ENST00000399002.7
ENST00000434210.5 |
SPTLC3
|
serine palmitoyltransferase long chain base subunit 3 |
| chr9_+_69820799 | 1.03 |
ENST00000377197.8
|
C9orf135
|
chromosome 9 open reading frame 135 |
| chr4_+_122379437 | 1.03 |
ENST00000439307.5
ENST00000388724.6 |
ADAD1
|
adenosine deaminase domain containing 1 |
| chr17_-_66220630 | 1.02 |
ENST00000585162.1
|
APOH
|
apolipoprotein H |
| chr4_+_70334963 | 1.02 |
ENST00000273936.6
|
CABS1
|
calcium binding protein, spermatid associated 1 |
| chr9_+_122159886 | 1.01 |
ENST00000373764.8
ENST00000536616.5 |
MORN5
|
MORN repeat containing 5 |
| chr10_+_15043937 | 1.01 |
ENST00000378228.8
ENST00000378217.3 |
OLAH
|
oleoyl-ACP hydrolase |
| chr3_+_186666003 | 1.01 |
ENST00000232003.5
|
HRG
|
histidine rich glycoprotein |
| chr11_-_85686123 | 1.00 |
ENST00000316398.5
|
CCDC89
|
coiled-coil domain containing 89 |
| chr1_-_159714581 | 0.99 |
ENST00000255030.9
ENST00000437342.1 ENST00000368112.5 ENST00000368111.5 ENST00000368110.1 |
CRP
|
C-reactive protein |
| chr9_-_34895722 | 0.99 |
ENST00000603640.6
ENST00000603592.1 ENST00000340783.11 |
FAM205C
|
family with sequence similarity 205 member C |
| chr4_+_122379508 | 0.98 |
ENST00000388725.2
|
ADAD1
|
adenosine deaminase domain containing 1 |
| chrX_+_30243715 | 0.98 |
ENST00000378981.8
ENST00000397550.6 |
MAGEB1
|
MAGE family member B1 |
| chr12_-_9999176 | 0.97 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chrX_+_9912548 | 0.97 |
ENST00000452575.1
|
SHROOM2
|
shroom family member 2 |
| chr9_-_97938157 | 0.97 |
ENST00000616898.2
|
HEMGN
|
hemogen |
| chr6_-_49866453 | 0.96 |
ENST00000507853.5
|
CRISP1
|
cysteine rich secretory protein 1 |
| chr1_+_103655760 | 0.95 |
ENST00000370083.9
|
AMY1A
|
amylase alpha 1A |
| chr10_-_96271508 | 0.95 |
ENST00000427367.6
ENST00000413476.6 ENST00000371176.6 |
BLNK
|
B cell linker |
| chr14_-_22819721 | 0.95 |
ENST00000554517.5
ENST00000285850.11 ENST00000397529.6 ENST00000555702.5 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr16_+_4734278 | 0.94 |
ENST00000586724.5
|
C16orf71
|
chromosome 16 open reading frame 71 |
| chr15_-_58014097 | 0.94 |
ENST00000559517.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr9_+_114323098 | 0.94 |
ENST00000259396.9
|
ORM1
|
orosomucoid 1 |
| chr11_+_86395166 | 0.93 |
ENST00000528728.1
|
CCDC81
|
coiled-coil domain containing 81 |
| chr2_+_200440649 | 0.92 |
ENST00000366118.2
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr19_-_32869741 | 0.91 |
ENST00000590341.5
ENST00000587772.1 ENST00000023064.9 |
SLC7A9
|
solute carrier family 7 member 9 |
| chr20_-_57359491 | 0.90 |
ENST00000543500.3
|
MTRNR2L3
|
MT-RNR2 like 3 |
| chr6_-_116060859 | 0.89 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr12_-_102197827 | 0.89 |
ENST00000329406.5
|
PMCH
|
pro-melanin concentrating hormone |
| chr3_-_134250831 | 0.89 |
ENST00000623711.4
|
RYK
|
receptor like tyrosine kinase |
| chr7_+_130207847 | 0.89 |
ENST00000297819.4
|
SSMEM1
|
serine rich single-pass membrane protein 1 |
| chr12_-_101739426 | 0.89 |
ENST00000266743.6
ENST00000392924.2 ENST00000392927.7 |
SYCP3
|
synaptonemal complex protein 3 |
| chr3_-_19934189 | 0.88 |
ENST00000295824.14
|
EFHB
|
EF-hand domain family member B |
| chr17_-_17582417 | 0.88 |
ENST00000395783.5
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr22_+_30356620 | 0.87 |
ENST00000399824.6
ENST00000405659.5 |
CCDC157
|
coiled-coil domain containing 157 |
| chr14_+_94581880 | 0.87 |
ENST00000557598.1
ENST00000556064.1 |
SERPINA5
|
serpin family A member 5 |
| chr4_-_184217854 | 0.86 |
ENST00000296741.7
|
ENPP6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr2_-_187554351 | 0.85 |
ENST00000437725.5
ENST00000409676.5 ENST00000233156.9 ENST00000339091.8 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor |
| chr19_-_4338786 | 0.85 |
ENST00000601482.1
ENST00000600324.5 ENST00000594605.6 |
STAP2
|
signal transducing adaptor family member 2 |
| chr7_-_73578657 | 0.85 |
ENST00000479892.5
|
TBL2
|
transducin beta like 2 |
| chr6_-_32666648 | 0.84 |
ENST00000399082.7
ENST00000399079.7 ENST00000374943.8 ENST00000434651.6 |
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr14_+_75632610 | 0.82 |
ENST00000555027.1
|
FLVCR2
|
FLVCR heme transporter 2 |
| chr2_+_79025709 | 0.82 |
ENST00000409471.1
|
REG3G
|
regenerating family member 3 gamma |
| chr2_-_105438503 | 0.82 |
ENST00000393352.7
ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr2_+_196713117 | 0.81 |
ENST00000409270.5
|
CCDC150
|
coiled-coil domain containing 150 |
| chr8_-_48921735 | 0.81 |
ENST00000396822.6
|
SNAI2
|
snail family transcriptional repressor 2 |
| chr1_+_248197265 | 0.80 |
ENST00000641626.1
|
OR2M3
|
olfactory receptor family 2 subfamily M member 3 |
| chr9_-_72060605 | 0.79 |
ENST00000377024.8
ENST00000651200.2 ENST00000652752.1 |
C9orf57
|
chromosome 9 open reading frame 57 |
| chr11_+_58943249 | 0.78 |
ENST00000300079.9
|
GLYATL1
|
glycine-N-acyltransferase like 1 |
| chr1_+_103749898 | 0.78 |
ENST00000622339.5
|
AMY1C
|
amylase alpha 1C |
| chr9_-_121050264 | 0.77 |
ENST00000223642.3
|
C5
|
complement C5 |
| chr11_+_102112445 | 0.77 |
ENST00000524575.5
|
YAP1
|
Yes1 associated transcriptional regulator |
| chr4_-_139280179 | 0.77 |
ENST00000398955.2
|
MGARP
|
mitochondria localized glutamic acid rich protein |
| chr5_+_122129533 | 0.77 |
ENST00000296600.5
ENST00000504912.1 ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chr18_-_33136075 | 0.76 |
ENST00000581852.5
|
CCDC178
|
coiled-coil domain containing 178 |
| chrX_+_139530730 | 0.76 |
ENST00000218099.7
|
F9
|
coagulation factor IX |
| chr3_-_167474026 | 0.75 |
ENST00000466903.1
ENST00000264677.8 |
SERPINI2
|
serpin family I member 2 |
| chr11_+_60396435 | 0.75 |
ENST00000395005.6
|
MS4A14
|
membrane spanning 4-domains A14 |
| chrX_+_88747225 | 0.73 |
ENST00000276127.9
ENST00000373111.5 |
CPXCR1
|
CPX chromosome region candidate 1 |
| chr2_-_156342348 | 0.72 |
ENST00000409572.5
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2 |
| chr2_-_179746040 | 0.72 |
ENST00000409343.5
|
ZNF385B
|
zinc finger protein 385B |
| chr4_+_74308463 | 0.72 |
ENST00000413830.6
|
EPGN
|
epithelial mitogen |
| chr10_-_73358853 | 0.72 |
ENST00000355577.8
ENST00000310715.7 |
CFAP70
|
cilia and flagella associated protein 70 |
| chr14_+_104801082 | 0.72 |
ENST00000342537.8
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr13_-_38990824 | 0.71 |
ENST00000379631.9
|
STOML3
|
stomatin like 3 |
| chr12_+_19205294 | 0.71 |
ENST00000424268.5
|
PLEKHA5
|
pleckstrin homology domain containing A5 |
| chr5_+_38403535 | 0.71 |
ENST00000336740.10
ENST00000397202.6 |
EGFLAM
|
EGF like, fibronectin type III and laminin G domains |
| chr12_+_7304284 | 0.71 |
ENST00000399422.4
|
ACSM4
|
acyl-CoA synthetase medium chain family member 4 |
| chr12_-_50396601 | 0.70 |
ENST00000327337.6
ENST00000543111.5 |
FAM186A
|
family with sequence similarity 186 member A |
| chr10_-_62236112 | 0.70 |
ENST00000315289.6
|
RTKN2
|
rhotekin 2 |
| chrX_-_70908705 | 0.69 |
ENST00000374333.7
ENST00000395889.6 |
TEX11
|
testis expressed 11 |
| chr14_+_96482982 | 0.69 |
ENST00000554706.1
|
AK7
|
adenylate kinase 7 |
| chr1_+_196977550 | 0.68 |
ENST00000256785.5
|
CFHR5
|
complement factor H related 5 |
| chr12_-_120325936 | 0.68 |
ENST00000549767.1
|
PLA2G1B
|
phospholipase A2 group IB |
| chr5_+_73626158 | 0.67 |
ENST00000296794.10
ENST00000545377.5 ENST00000509848.5 ENST00000513042.7 |
ARHGEF28
|
Rho guanine nucleotide exchange factor 28 |
| chr2_-_224569782 | 0.67 |
ENST00000409096.5
|
CUL3
|
cullin 3 |
| chrX_+_139530752 | 0.67 |
ENST00000394090.2
|
F9
|
coagulation factor IX |
| chr15_-_51243011 | 0.66 |
ENST00000405913.7
ENST00000559878.5 |
CYP19A1
|
cytochrome P450 family 19 subfamily A member 1 |
| chr6_-_28587250 | 0.66 |
ENST00000452236.3
|
ZBED9
|
zinc finger BED-type containing 9 |
| chr3_-_49029378 | 0.66 |
ENST00000442157.2
ENST00000326739.9 ENST00000677010.1 ENST00000678724.1 ENST00000429182.6 |
IMPDH2
|
inosine monophosphate dehydrogenase 2 |
| chrX_-_100636799 | 0.65 |
ENST00000373020.9
|
TSPAN6
|
tetraspanin 6 |
| chr13_-_38990856 | 0.65 |
ENST00000423210.1
|
STOML3
|
stomatin like 3 |
| chr2_+_21137468 | 0.65 |
ENST00000622654.1
|
TDRD15
|
tudor domain containing 15 |
| chr11_+_22666604 | 0.65 |
ENST00000454584.6
|
GAS2
|
growth arrest specific 2 |
| chr6_+_38715288 | 0.65 |
ENST00000327475.11
|
DNAH8
|
dynein axonemal heavy chain 8 |
| chr4_-_110199173 | 0.64 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr1_+_172452885 | 0.64 |
ENST00000367725.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr10_+_5048748 | 0.64 |
ENST00000602997.5
ENST00000439082.7 |
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr7_+_1232865 | 0.64 |
ENST00000316333.9
|
UNCX
|
UNC homeobox |
| chr4_-_110198976 | 0.64 |
ENST00000506625.5
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr4_-_110198650 | 0.63 |
ENST00000394607.7
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr3_+_184812385 | 0.63 |
ENST00000453056.5
|
VPS8
|
VPS8 subunit of CORVET complex |
| chr6_-_25830557 | 0.63 |
ENST00000468082.1
|
SLC17A1
|
solute carrier family 17 member 1 |
| chr15_-_56465130 | 0.63 |
ENST00000260453.4
|
MNS1
|
meiosis specific nuclear structural 1 |
| chrX_-_143721423 | 0.62 |
ENST00000598475.1
|
SPANXN2
|
SPANX family member N2 |
| chr2_+_142877653 | 0.62 |
ENST00000375773.6
ENST00000409512.5 ENST00000264170.9 ENST00000410015.6 |
KYNU
|
kynureninase |
| chr3_-_57544324 | 0.61 |
ENST00000495027.5
ENST00000351747.6 ENST00000389536.8 ENST00000311202.7 |
DNAH12
|
dynein axonemal heavy chain 12 |
| chr19_+_14028148 | 0.61 |
ENST00000431365.3
ENST00000585987.1 |
RLN3
|
relaxin 3 |
| chr13_-_99258366 | 0.61 |
ENST00000397470.5
ENST00000397473.7 |
GPR18
|
G protein-coupled receptor 18 |
| chr10_-_72954790 | 0.60 |
ENST00000373032.4
|
PLA2G12B
|
phospholipase A2 group XIIB |
| chr6_+_135851681 | 0.60 |
ENST00000308191.11
|
PDE7B
|
phosphodiesterase 7B |
| chr6_-_29005313 | 0.60 |
ENST00000377179.4
|
ZNF311
|
zinc finger protein 311 |
| chr16_+_77199408 | 0.60 |
ENST00000378644.5
|
SYCE1L
|
synaptonemal complex central element protein 1 like |
| chr1_-_56579555 | 0.58 |
ENST00000371250.4
|
PLPP3
|
phospholipid phosphatase 3 |
| chr10_+_94762673 | 0.57 |
ENST00000480405.2
ENST00000371321.9 |
CYP2C19
|
cytochrome P450 family 2 subfamily C member 19 |
| chr6_-_134950081 | 0.57 |
ENST00000367847.2
ENST00000265605.7 ENST00000367845.6 |
ALDH8A1
|
aldehyde dehydrogenase 8 family member A1 |
| chr19_-_58353482 | 0.56 |
ENST00000263100.8
|
A1BG
|
alpha-1-B glycoprotein |
| chr8_-_11996310 | 0.56 |
ENST00000526438.5
ENST00000382205.4 |
DEFB134
|
defensin beta 134 |
| chr3_+_194136138 | 0.56 |
ENST00000232424.4
|
HES1
|
hes family bHLH transcription factor 1 |
| chr17_-_48613468 | 0.56 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr20_+_31739260 | 0.55 |
ENST00000340513.4
ENST00000300403.11 |
TPX2
|
TPX2 microtubule nucleation factor |
| chrX_-_13319952 | 0.55 |
ENST00000622204.1
ENST00000380622.5 |
ATXN3L
|
ataxin 3 like |
| chr10_+_35195843 | 0.55 |
ENST00000488741.5
ENST00000474931.5 ENST00000468236.5 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr14_-_76826229 | 0.54 |
ENST00000557497.1
|
ANGEL1
|
angel homolog 1 |
| chrX_+_630789 | 0.54 |
ENST00000381575.6
|
SHOX
|
short stature homeobox |
| chr5_+_73813518 | 0.54 |
ENST00000296799.8
|
ARHGEF28
|
Rho guanine nucleotide exchange factor 28 |
| chr4_+_95051671 | 0.54 |
ENST00000440890.7
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr10_+_99782628 | 0.53 |
ENST00000648689.1
ENST00000647814.1 |
ABCC2
|
ATP binding cassette subfamily C member 2 |
| chr12_+_19205257 | 0.53 |
ENST00000538305.5
|
PLEKHA5
|
pleckstrin homology domain containing A5 |
| chr4_-_69760610 | 0.53 |
ENST00000310613.8
|
SULT1B1
|
sulfotransferase family 1B member 1 |
| chr6_+_147508645 | 0.53 |
ENST00000367474.2
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr17_-_35930727 | 0.53 |
ENST00000616596.4
ENST00000612980.4 ENST00000613308.4 ENST00000619876.4 |
RDM1
|
RAD52 motif containing 1 |
| chr17_-_40417873 | 0.52 |
ENST00000423485.6
|
TOP2A
|
DNA topoisomerase II alpha |
| chr20_-_63956382 | 0.52 |
ENST00000358711.7
ENST00000354216.11 |
UCKL1
|
uridine-cytidine kinase 1 like 1 |
| chr16_-_65072207 | 0.52 |
ENST00000562882.5
ENST00000567934.5 |
CDH11
|
cadherin 11 |
| chr10_-_52772763 | 0.52 |
ENST00000675947.1
ENST00000674931.1 |
MBL2
|
mannose binding lectin 2 |
| chr9_-_35812142 | 0.52 |
ENST00000497810.1
ENST00000484764.5 |
SPAG8
|
sperm associated antigen 8 |
| chr7_+_127887912 | 0.51 |
ENST00000486037.1
|
SND1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr10_+_21524670 | 0.51 |
ENST00000631589.1
|
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chr17_-_41140487 | 0.51 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6 |
| chr9_-_35563867 | 0.51 |
ENST00000399742.7
ENST00000619051.4 |
FAM166B
|
family with sequence similarity 166 member B |
| chr1_-_53940100 | 0.51 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr5_-_111756245 | 0.50 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr4_-_80073170 | 0.50 |
ENST00000403729.7
|
ANTXR2
|
ANTXR cell adhesion molecule 2 |
| chr12_-_54188871 | 0.50 |
ENST00000504338.5
ENST00000514685.5 ENST00000504797.1 ENST00000513838.5 ENST00000505128.5 ENST00000337581.7 ENST00000503306.5 ENST00000243112.9 ENST00000514196.5 ENST00000682136.1 ENST00000506169.5 ENST00000507904.5 ENST00000508394.6 |
SMUG1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| chr2_+_234050679 | 0.50 |
ENST00000373368.5
ENST00000168148.8 |
SPP2
|
secreted phosphoprotein 2 |
| chr10_+_15043919 | 0.50 |
ENST00000378225.5
|
OLAH
|
oleoyl-ACP hydrolase |
| chr12_+_106357729 | 0.50 |
ENST00000228347.9
|
POLR3B
|
RNA polymerase III subunit B |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXQ1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.8 | 2.4 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.7 | 2.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.7 | 2.2 | GO:0060721 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.7 | 2.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.7 | 2.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.6 | 2.5 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.5 | 1.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.4 | 1.3 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.4 | 1.2 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.4 | 1.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.3 | 0.6 | GO:2000974 | inhibition of neuroepithelial cell differentiation(GO:0002085) negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.3 | 0.8 | GO:0046247 | tetraterpenoid metabolic process(GO:0016108) carotenoid metabolic process(GO:0016116) carotene catabolic process(GO:0016121) xanthophyll metabolic process(GO:0016122) terpene catabolic process(GO:0046247) |
| 0.3 | 9.2 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.3 | 1.6 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.3 | 1.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.3 | 1.0 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.2 | 2.5 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 0.5 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.2 | 0.6 | GO:0016487 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.2 | 0.6 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.2 | 2.9 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 0.6 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.2 | 0.7 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.2 | 0.5 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.2 | 2.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 1.5 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 0.8 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.2 | 1.2 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 2.1 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.9 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.1 | 0.9 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 0.7 | GO:0006311 | meiotic gene conversion(GO:0006311) |
| 0.1 | 0.7 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.5 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.4 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.1 | 1.5 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.3 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.3 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.9 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.8 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.9 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.5 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) regulation of melanosome transport(GO:1902908) |
| 0.1 | 0.9 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.9 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 2.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.5 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.5 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 1.0 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.1 | 1.2 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 | 0.3 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 1.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.3 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 0.6 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.1 | 0.6 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.4 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 1.4 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 2.0 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 0.2 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 2.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.3 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.1 | 1.0 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.9 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 1.0 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.9 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.1 | 1.9 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.5 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.5 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.8 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 0.3 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.3 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.4 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 0.4 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.5 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.8 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.2 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.3 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 1.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.2 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 1.0 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 1.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.8 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 1.0 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 | 0.6 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.2 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.4 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.3 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.2 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.4 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.9 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 2.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.4 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 3.7 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.3 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.6 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.2 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.7 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.0 | 0.8 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.5 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.2 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.6 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.5 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 0.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0000820 | regulation of glutamine family amino acid metabolic process(GO:0000820) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 1.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.4 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.4 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.4 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.2 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.6 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 1.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.2 | GO:0070836 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.4 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.6 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.6 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.0 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 2.9 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.2 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.0 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) female genitalia morphogenesis(GO:0048807) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.0 | 0.3 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.6 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.7 | GO:0002381 | immunoglobulin production involved in immunoglobulin mediated immune response(GO:0002381) |
| 0.0 | 0.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 2.0 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.2 | 0.9 | GO:0097182 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.2 | 2.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 0.8 | GO:0071149 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.7 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 1.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.8 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 2.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.7 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.3 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 7.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 4.0 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.9 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.3 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 1.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.5 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.5 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 1.0 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) translation initiation ternary complex(GO:0044207) |
| 0.0 | 0.6 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.0 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 2.2 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.4 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 2.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 3.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.8 | GO:0031430 | M band(GO:0031430) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.4 | 1.2 | GO:0015489 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.3 | 1.3 | GO:0033265 | choline binding(GO:0033265) |
| 0.3 | 1.5 | GO:0016295 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.3 | 0.9 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.3 | 1.0 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.2 | 2.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 1.5 | GO:0019862 | IgA binding(GO:0019862) |
| 0.2 | 0.6 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.2 | 0.8 | GO:0047946 | glutamine N-acyltransferase activity(GO:0047946) |
| 0.2 | 0.5 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 1.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 3.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.2 | 0.5 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.2 | 9.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 0.8 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 2.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 2.5 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 2.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.5 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.6 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.5 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 3.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.4 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.1 | 0.6 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.5 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.9 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.5 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.6 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.9 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.3 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 0.7 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 1.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 2.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.7 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.9 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 1.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.4 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 1.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.9 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 1.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.2 | GO:0003860 | 3-hydroxyisobutyryl-CoA hydrolase activity(GO:0003860) |
| 0.1 | 0.4 | GO:0005119 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.9 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 1.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 1.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.5 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 1.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.3 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 2.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 2.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 1.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 3.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0043125 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 2.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 7.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 2.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 2.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 6.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 9.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 2.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 2.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 3.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.5 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 1.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 2.0 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |