Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
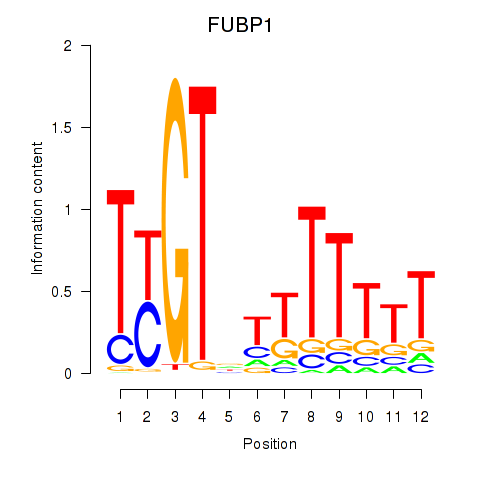
Results for FUBP1
Z-value: 1.39
Transcription factors associated with FUBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FUBP1
|
ENSG00000162613.17 | far upstream element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FUBP1 | hg38_v1_chr1_-_77979054_77979116 | -0.19 | 3.0e-01 | Click! |
Activity profile of FUBP1 motif
Sorted Z-values of FUBP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_10568650 | 6.24 |
ENST00000256178.8
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr11_-_10568571 | 6.18 |
ENST00000529598.1
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr11_-_107719657 | 5.45 |
ENST00000525934.1
ENST00000531293.1 |
SLN
|
sarcolipin |
| chr7_+_116526277 | 5.34 |
ENST00000393468.1
ENST00000393467.1 |
CAV1
|
caveolin 1 |
| chr5_+_40909490 | 4.96 |
ENST00000313164.10
|
C7
|
complement C7 |
| chr7_+_80638510 | 4.88 |
ENST00000433696.6
ENST00000538969.5 ENST00000544133.5 |
CD36
|
CD36 molecule |
| chr12_-_91182784 | 4.87 |
ENST00000547568.6
ENST00000052754.10 ENST00000552962.5 |
DCN
|
decorin |
| chr7_+_80638633 | 4.65 |
ENST00000447544.7
ENST00000482059.6 |
CD36
|
CD36 molecule |
| chr4_-_151226427 | 4.36 |
ENST00000304527.8
ENST00000409598.8 |
SH3D19
|
SH3 domain containing 19 |
| chr12_-_91182652 | 4.30 |
ENST00000552145.5
ENST00000546745.5 |
DCN
|
decorin |
| chr1_+_43300971 | 4.28 |
ENST00000372476.8
ENST00000538015.1 |
TIE1
|
tyrosine kinase with immunoglobulin like and EGF like domains 1 |
| chr12_-_110920568 | 4.08 |
ENST00000548438.1
ENST00000228841.15 |
MYL2
|
myosin light chain 2 |
| chr17_-_10518536 | 4.00 |
ENST00000226207.6
|
MYH1
|
myosin heavy chain 1 |
| chr1_-_27374816 | 3.98 |
ENST00000270879.9
ENST00000354982.2 |
FCN3
|
ficolin 3 |
| chr3_+_29281552 | 3.92 |
ENST00000452462.5
ENST00000456853.1 |
RBMS3
|
RNA binding motif single stranded interacting protein 3 |
| chr10_+_61901678 | 3.64 |
ENST00000644638.1
ENST00000681100.1 ENST00000279873.12 |
ARID5B
|
AT-rich interaction domain 5B |
| chr17_-_43661915 | 3.48 |
ENST00000318579.9
ENST00000393661.2 |
MEOX1
|
mesenchyme homeobox 1 |
| chr19_-_49362376 | 3.41 |
ENST00000601519.5
ENST00000593945.6 ENST00000539846.5 ENST00000596757.1 ENST00000311227.6 |
TEAD2
|
TEA domain transcription factor 2 |
| chr12_+_53973108 | 3.41 |
ENST00000546378.1
ENST00000243082.4 |
HOXC11
|
homeobox C11 |
| chr4_-_100517991 | 3.16 |
ENST00000511970.5
ENST00000502569.1 ENST00000305864.7 ENST00000296420.9 |
EMCN
|
endomucin |
| chr1_+_202348687 | 3.14 |
ENST00000608999.6
ENST00000391959.5 ENST00000480184.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr1_+_77918128 | 3.00 |
ENST00000342754.5
|
NEXN
|
nexilin F-actin binding protein |
| chrX_-_54994022 | 2.81 |
ENST00000614686.1
ENST00000374992.6 ENST00000375006.8 |
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr15_-_55917129 | 2.70 |
ENST00000338963.6
ENST00000508342.5 |
NEDD4
|
NEDD4 E3 ubiquitin protein ligase |
| chr3_-_187736493 | 2.63 |
ENST00000232014.8
|
BCL6
|
BCL6 transcription repressor |
| chr17_-_64390852 | 2.57 |
ENST00000563924.6
|
PECAM1
|
platelet and endothelial cell adhesion molecule 1 |
| chr4_-_16898619 | 2.50 |
ENST00000502640.5
ENST00000304523.10 ENST00000506732.1 |
LDB2
|
LIM domain binding 2 |
| chr1_+_31413806 | 2.47 |
ENST00000536384.2
|
SERINC2
|
serine incorporator 2 |
| chr9_-_107489754 | 2.40 |
ENST00000610832.1
ENST00000374672.5 |
KLF4
|
Kruppel like factor 4 |
| chr15_-_55917080 | 2.38 |
ENST00000506154.1
|
NEDD4
|
NEDD4 E3 ubiquitin protein ligase |
| chr11_+_75562242 | 2.32 |
ENST00000526397.5
ENST00000529643.1 ENST00000525492.5 ENST00000530284.5 ENST00000358171.8 |
SERPINH1
|
serpin family H member 1 |
| chr17_-_64390592 | 2.27 |
ENST00000563523.5
|
PECAM1
|
platelet and endothelial cell adhesion molecule 1 |
| chr1_-_146021724 | 2.26 |
ENST00000475797.1
ENST00000497365.5 ENST00000336751.11 ENST00000634927.1 ENST00000421822.2 |
HJV
|
hemojuvelin BMP co-receptor |
| chr11_+_60914139 | 2.25 |
ENST00000227525.8
|
TMEM109
|
transmembrane protein 109 |
| chr11_+_75562274 | 2.24 |
ENST00000532356.5
ENST00000524558.5 |
SERPINH1
|
serpin family H member 1 |
| chr11_+_114060204 | 2.16 |
ENST00000683318.1
|
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chr17_-_43661563 | 2.11 |
ENST00000549132.2
|
MEOX1
|
mesenchyme homeobox 1 |
| chr17_+_9021501 | 2.00 |
ENST00000173229.7
|
NTN1
|
netrin 1 |
| chr2_-_178804623 | 1.98 |
ENST00000359218.10
ENST00000342175.11 |
TTN
|
titin |
| chr19_-_45493208 | 1.96 |
ENST00000430715.6
|
RTN2
|
reticulon 2 |
| chr4_-_16898561 | 1.95 |
ENST00000515064.5
ENST00000441778.6 |
LDB2
|
LIM domain binding 2 |
| chr4_-_151227881 | 1.94 |
ENST00000652233.1
ENST00000514152.5 |
SH3D19
|
SH3 domain containing 19 |
| chr3_-_149333619 | 1.91 |
ENST00000296059.7
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr18_-_55588535 | 1.87 |
ENST00000566286.5
ENST00000566279.5 ENST00000626595.2 ENST00000564999.5 ENST00000616053.4 ENST00000356073.8 |
TCF4
|
transcription factor 4 |
| chr15_-_29822418 | 1.85 |
ENST00000614355.5
ENST00000495972.6 ENST00000346128.10 |
TJP1
|
tight junction protein 1 |
| chr16_+_28553908 | 1.82 |
ENST00000317058.8
|
SGF29
|
SAGA complex associated factor 29 |
| chr2_-_72147819 | 1.80 |
ENST00000001146.7
ENST00000546307.5 ENST00000474509.1 |
CYP26B1
|
cytochrome P450 family 26 subfamily B member 1 |
| chr18_+_7754959 | 1.79 |
ENST00000400053.8
|
PTPRM
|
protein tyrosine phosphatase receptor type M |
| chr22_-_38317380 | 1.75 |
ENST00000413574.6
|
CSNK1E
|
casein kinase 1 epsilon |
| chrX_+_96684638 | 1.74 |
ENST00000355827.8
ENST00000373061.7 |
DIAPH2
|
diaphanous related formin 2 |
| chrX_-_66040072 | 1.70 |
ENST00000374737.9
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chrX_-_33128360 | 1.68 |
ENST00000378677.6
|
DMD
|
dystrophin |
| chr12_-_85836372 | 1.67 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9 |
| chr4_+_81030700 | 1.60 |
ENST00000282701.4
|
BMP3
|
bone morphogenetic protein 3 |
| chrX_-_66040057 | 1.57 |
ENST00000412866.2
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr9_+_19409000 | 1.55 |
ENST00000340967.3
|
ACER2
|
alkaline ceramidase 2 |
| chr12_-_21941300 | 1.55 |
ENST00000684084.1
|
ABCC9
|
ATP binding cassette subfamily C member 9 |
| chrX_-_66040107 | 1.54 |
ENST00000455586.6
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr12_+_48119787 | 1.54 |
ENST00000551804.5
|
PFKM
|
phosphofructokinase, muscle |
| chr12_-_21941402 | 1.54 |
ENST00000326684.8
ENST00000682068.1 ENST00000621589.2 ENST00000261200.9 ENST00000683676.1 |
ABCC9
|
ATP binding cassette subfamily C member 9 |
| chr9_-_127877665 | 1.50 |
ENST00000644144.2
|
AK1
|
adenylate kinase 1 |
| chr6_-_56642788 | 1.41 |
ENST00000439203.5
ENST00000518935.5 ENST00000370765.11 ENST00000244364.10 |
DST
|
dystonin |
| chrX_-_154353168 | 1.40 |
ENST00000444578.1
|
FLNA
|
filamin A |
| chr12_+_53990585 | 1.36 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr4_-_44651619 | 1.34 |
ENST00000415895.9
ENST00000332990.6 |
YIPF7
|
Yip1 domain family member 7 |
| chr12_-_9869345 | 1.31 |
ENST00000228438.3
|
CLEC2B
|
C-type lectin domain family 2 member B |
| chr10_-_104085847 | 1.28 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr11_+_7485606 | 1.27 |
ENST00000528758.1
|
OLFML1
|
olfactomedin like 1 |
| chr18_-_55588184 | 1.27 |
ENST00000354452.8
ENST00000565908.6 ENST00000635822.2 |
TCF4
|
transcription factor 4 |
| chr20_+_9069076 | 1.25 |
ENST00000378473.9
|
PLCB4
|
phospholipase C beta 4 |
| chr22_-_38316973 | 1.25 |
ENST00000430335.1
|
CSNK1E
|
casein kinase 1 epsilon |
| chrX_+_96684712 | 1.24 |
ENST00000373049.8
|
DIAPH2
|
diaphanous related formin 2 |
| chr19_+_13024573 | 1.24 |
ENST00000358552.7
ENST00000360105.8 ENST00000588228.5 ENST00000676441.1 ENST00000591028.1 |
NFIX
|
nuclear factor I X |
| chr7_+_120989030 | 1.20 |
ENST00000428526.5
|
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr5_+_145936554 | 1.18 |
ENST00000359120.9
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr6_-_111567271 | 1.18 |
ENST00000464903.1
ENST00000368730.5 ENST00000368735.1 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr11_+_110130992 | 1.17 |
ENST00000528673.5
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chrX_+_38352573 | 1.17 |
ENST00000039007.5
|
OTC
|
ornithine transcarbamylase |
| chr17_-_76053639 | 1.13 |
ENST00000602720.5
|
SRP68
|
signal recognition particle 68 |
| chr6_+_121435595 | 1.13 |
ENST00000649003.1
ENST00000282561.4 |
GJA1
|
gap junction protein alpha 1 |
| chr4_-_185657520 | 1.11 |
ENST00000438278.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr12_-_8650529 | 1.11 |
ENST00000543467.5
|
MFAP5
|
microfibril associated protein 5 |
| chr3_-_149333407 | 1.11 |
ENST00000470080.5
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr2_-_65366650 | 1.09 |
ENST00000443619.6
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chr10_-_104085793 | 1.06 |
ENST00000650263.1
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr14_+_61187544 | 1.06 |
ENST00000555185.5
ENST00000557294.5 ENST00000556778.5 |
PRKCH
|
protein kinase C eta |
| chr6_+_32164586 | 1.05 |
ENST00000333845.11
ENST00000395512.5 ENST00000432129.1 |
EGFL8
|
EGF like domain multiple 8 |
| chr12_+_25958891 | 1.03 |
ENST00000381352.7
ENST00000535907.5 ENST00000405154.6 ENST00000615708.4 |
RASSF8
|
Ras association domain family member 8 |
| chr19_+_44751251 | 1.02 |
ENST00000444487.1
|
BCL3
|
BCL3 transcription coactivator |
| chr2_+_161231078 | 1.00 |
ENST00000439442.1
|
TANK
|
TRAF family member associated NFKB activator |
| chr19_+_1438351 | 1.00 |
ENST00000233609.8
|
RPS15
|
ribosomal protein S15 |
| chr22_+_37024137 | 0.96 |
ENST00000628507.1
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr10_-_104085762 | 0.95 |
ENST00000393211.3
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr2_+_176151543 | 0.91 |
ENST00000306324.4
|
HOXD4
|
homeobox D4 |
| chr1_-_11681096 | 0.90 |
ENST00000445656.5
ENST00000376669.9 ENST00000456915.1 ENST00000376692.9 |
MAD2L2
|
mitotic arrest deficient 2 like 2 |
| chr2_+_147844601 | 0.88 |
ENST00000404590.1
|
ACVR2A
|
activin A receptor type 2A |
| chr10_+_24042131 | 0.86 |
ENST00000636305.1
|
KIAA1217
|
KIAA1217 |
| chr12_-_91058016 | 0.84 |
ENST00000266719.4
|
KERA
|
keratocan |
| chr21_-_37073019 | 0.81 |
ENST00000360525.9
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis class P |
| chr19_+_18571730 | 0.81 |
ENST00000596304.5
ENST00000430157.6 ENST00000596273.5 ENST00000442744.7 ENST00000595683.5 ENST00000599256.5 ENST00000595158.5 ENST00000598780.5 |
UBA52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr2_+_201116143 | 0.78 |
ENST00000443227.5
ENST00000309955.8 ENST00000341222.10 ENST00000341582.10 |
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr6_+_125203639 | 0.78 |
ENST00000392482.6
|
TPD52L1
|
TPD52 like 1 |
| chr2_+_147844488 | 0.77 |
ENST00000535787.5
|
ACVR2A
|
activin A receptor type 2A |
| chr6_-_31645025 | 0.75 |
ENST00000438149.5
|
BAG6
|
BAG cochaperone 6 |
| chr7_-_22193824 | 0.75 |
ENST00000401957.6
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr11_-_47499757 | 0.72 |
ENST00000535982.1
|
CELF1
|
CUGBP Elav-like family member 1 |
| chr2_+_33134620 | 0.72 |
ENST00000402934.5
ENST00000404525.5 ENST00000407925.5 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr12_-_68225806 | 0.70 |
ENST00000229134.5
|
IL26
|
interleukin 26 |
| chr19_+_13023958 | 0.70 |
ENST00000587760.5
ENST00000585575.5 |
NFIX
|
nuclear factor I X |
| chr17_+_28719995 | 0.68 |
ENST00000496182.5
|
RPL23A
|
ribosomal protein L23a |
| chr3_+_130440510 | 0.68 |
ENST00000373157.8
|
COL6A5
|
collagen type VI alpha 5 chain |
| chr6_+_5260992 | 0.68 |
ENST00000324331.10
|
FARS2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr18_+_63702958 | 0.67 |
ENST00000544088.6
|
SERPINB11
|
serpin family B member 11 |
| chr17_+_8247601 | 0.67 |
ENST00000585183.5
|
PFAS
|
phosphoribosylformylglycinamidine synthase |
| chr3_-_71064915 | 0.64 |
ENST00000614176.5
ENST00000485326.7 |
FOXP1
|
forkhead box P1 |
| chr19_-_14090963 | 0.63 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr3_+_148827800 | 0.61 |
ENST00000282957.9
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 |
| chr11_+_55723776 | 0.61 |
ENST00000641440.1
|
OR5D3P
|
olfactory receptor family 5 subfamily D member 3 pseudogene |
| chr21_-_37072688 | 0.59 |
ENST00000464265.5
ENST00000399102.5 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis class P |
| chr2_-_287687 | 0.58 |
ENST00000401489.6
ENST00000619265.4 |
ALKAL2
|
ALK and LTK ligand 2 |
| chr19_+_11435272 | 0.57 |
ENST00000676823.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr12_-_21941225 | 0.56 |
ENST00000538350.5
|
ABCC9
|
ATP binding cassette subfamily C member 9 |
| chr17_-_3691887 | 0.55 |
ENST00000552050.5
|
P2RX5
|
purinergic receptor P2X 5 |
| chr6_-_31645503 | 0.54 |
ENST00000453833.5
|
BAG6
|
BAG cochaperone 6 |
| chr7_+_75092573 | 0.53 |
ENST00000651028.1
ENST00000472837.7 ENST00000629105.2 ENST00000619142.4 ENST00000614064.4 |
GTF2IRD2B
|
GTF2I repeat domain containing 2B |
| chr18_-_63318674 | 0.53 |
ENST00000589955.2
|
BCL2
|
BCL2 apoptosis regulator |
| chr20_+_9069095 | 0.51 |
ENST00000437503.5
|
PLCB4
|
phospholipase C beta 4 |
| chr22_-_29553645 | 0.51 |
ENST00000397871.5
ENST00000397872.5 ENST00000440771.5 |
THOC5
|
THO complex 5 |
| chr18_-_55588146 | 0.51 |
ENST00000627784.2
|
TCF4
|
transcription factor 4 |
| chr2_+_33134579 | 0.50 |
ENST00000418533.6
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr3_+_57890011 | 0.50 |
ENST00000494088.6
ENST00000438794.5 |
SLMAP
|
sarcolemma associated protein |
| chr7_-_22194709 | 0.50 |
ENST00000458533.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr1_+_88684222 | 0.50 |
ENST00000316005.11
ENST00000370521.8 |
PKN2
|
protein kinase N2 |
| chr3_+_130560334 | 0.48 |
ENST00000358511.10
|
COL6A6
|
collagen type VI alpha 6 chain |
| chr10_-_54801179 | 0.47 |
ENST00000373955.5
|
PCDH15
|
protocadherin related 15 |
| chr4_-_83284752 | 0.47 |
ENST00000311461.7
ENST00000647002.2 ENST00000311469.9 |
COQ2
|
coenzyme Q2, polyprenyltransferase |
| chr19_+_40588706 | 0.47 |
ENST00000595726.1
|
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr22_-_50532137 | 0.46 |
ENST00000405135.5
ENST00000401779.5 ENST00000682240.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr1_-_154206327 | 0.44 |
ENST00000368525.4
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr2_-_10812706 | 0.44 |
ENST00000272227.8
|
PDIA6
|
protein disulfide isomerase family A member 6 |
| chr22_-_40856565 | 0.43 |
ENST00000620312.4
ENST00000216218.8 |
ST13
|
ST13 Hsp70 interacting protein |
| chr6_-_49969625 | 0.42 |
ENST00000398718.1
|
DEFB113
|
defensin beta 113 |
| chr1_+_202348727 | 0.41 |
ENST00000356764.6
|
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr7_+_130070518 | 0.41 |
ENST00000335420.10
ENST00000463413.1 |
KLHDC10
|
kelch domain containing 10 |
| chr6_+_144344101 | 0.41 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr1_+_44674728 | 0.40 |
ENST00000453711.1
|
ARMH1
|
armadillo like helical domain containing 1 |
| chr12_-_21941346 | 0.40 |
ENST00000684435.1
|
ABCC9
|
ATP binding cassette subfamily C member 9 |
| chr7_-_15561986 | 0.40 |
ENST00000342526.8
|
AGMO
|
alkylglycerol monooxygenase |
| chr2_+_147845020 | 0.39 |
ENST00000241416.12
|
ACVR2A
|
activin A receptor type 2A |
| chr13_-_51111209 | 0.39 |
ENST00000563710.6
|
C13orf42
|
chromosome 13 open reading frame 42 |
| chr11_+_55819630 | 0.37 |
ENST00000333976.7
|
OR5D18
|
olfactory receptor family 5 subfamily D member 18 |
| chr6_-_27867581 | 0.37 |
ENST00000331442.5
|
H1-5
|
H1.5 linker histone, cluster member |
| chr1_+_247605586 | 0.36 |
ENST00000320002.3
|
OR2G3
|
olfactory receptor family 2 subfamily G member 3 |
| chr1_+_43389889 | 0.36 |
ENST00000562955.2
ENST00000634258.3 |
SZT2
|
SZT2 subunit of KICSTOR complex |
| chr6_+_29111560 | 0.36 |
ENST00000377169.2
|
OR2J3
|
olfactory receptor family 2 subfamily J member 3 |
| chr10_-_54801221 | 0.36 |
ENST00000644397.2
ENST00000395445.6 ENST00000613657.5 ENST00000320301.11 |
PCDH15
|
protocadherin related 15 |
| chr2_+_33134406 | 0.35 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr19_+_15944299 | 0.35 |
ENST00000641275.1
|
OR10H4
|
olfactory receptor family 10 subfamily H member 4 |
| chr6_+_34757473 | 0.35 |
ENST00000244520.10
ENST00000374018.5 ENST00000374017.3 |
SNRPC
|
small nuclear ribonucleoprotein polypeptide C |
| chr3_+_48447093 | 0.34 |
ENST00000412052.4
|
ATRIP
|
ATR interacting protein |
| chr17_-_8248035 | 0.32 |
ENST00000651323.1
|
CTC1
|
CST telomere replication complex component 1 |
| chr8_-_20183090 | 0.31 |
ENST00000265808.11
ENST00000522513.5 |
SLC18A1
|
solute carrier family 18 member A1 |
| chr6_-_31541937 | 0.31 |
ENST00000456662.5
ENST00000431908.5 ENST00000456976.5 ENST00000428450.5 ENST00000418897.5 ENST00000396172.6 ENST00000419020.1 ENST00000428098.5 |
DDX39B
|
DExD-box helicase 39B |
| chr7_+_142111739 | 0.31 |
ENST00000550469.6
ENST00000477922.3 |
MGAM2
|
maltase-glucoamylase 2 (putative) |
| chr12_+_10212483 | 0.31 |
ENST00000545859.5
|
GABARAPL1
|
GABA type A receptor associated protein like 1 |
| chr2_+_180980807 | 0.31 |
ENST00000602475.5
|
UBE2E3
|
ubiquitin conjugating enzyme E2 E3 |
| chr11_+_1853049 | 0.30 |
ENST00000311604.8
|
LSP1
|
lymphocyte specific protein 1 |
| chr3_-_51499950 | 0.30 |
ENST00000423656.5
ENST00000504652.5 ENST00000684031.1 |
DCAF1
|
DDB1 and CUL4 associated factor 1 |
| chr8_+_69563824 | 0.30 |
ENST00000525999.5
|
SULF1
|
sulfatase 1 |
| chr11_+_57763820 | 0.29 |
ENST00000674106.1
|
CTNND1
|
catenin delta 1 |
| chr10_-_32378720 | 0.26 |
ENST00000375110.6
|
EPC1
|
enhancer of polycomb homolog 1 |
| chr1_+_156061142 | 0.26 |
ENST00000361084.10
|
RAB25
|
RAB25, member RAS oncogene family |
| chr6_-_5260883 | 0.24 |
ENST00000330636.9
|
LYRM4
|
LYR motif containing 4 |
| chr9_+_125748175 | 0.24 |
ENST00000491787.7
ENST00000447726.6 |
PBX3
|
PBX homeobox 3 |
| chr14_-_21383960 | 0.23 |
ENST00000555943.1
|
SUPT16H
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr12_+_15956585 | 0.23 |
ENST00000526530.1
|
DERA
|
deoxyribose-phosphate aldolase |
| chr5_+_157180816 | 0.23 |
ENST00000422843.8
|
ITK
|
IL2 inducible T cell kinase |
| chr4_-_118836067 | 0.23 |
ENST00000280551.11
|
SEC24D
|
SEC24 homolog D, COPII coat complex component |
| chr12_+_10212867 | 0.22 |
ENST00000545047.5
ENST00000266458.10 ENST00000629504.1 ENST00000543602.5 ENST00000545887.1 |
GABARAPL1
|
GABA type A receptor associated protein like 1 |
| chr19_+_19192229 | 0.21 |
ENST00000421262.7
ENST00000456252.7 ENST00000303088.9 ENST00000593273.5 |
RFXANK
|
regulatory factor X associated ankyrin containing protein |
| chr2_-_151261755 | 0.21 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr12_+_10212836 | 0.21 |
ENST00000421801.6
ENST00000544284.5 |
GABARAPL1
|
GABA type A receptor associated protein like 1 |
| chr11_-_5548540 | 0.20 |
ENST00000322653.7
|
OR52H1
|
olfactory receptor family 52 subfamily H member 1 |
| chr22_-_29553691 | 0.19 |
ENST00000490103.6
ENST00000397873.6 |
THOC5
|
THO complex 5 |
| chr1_-_247723764 | 0.18 |
ENST00000419891.1
|
ENSG00000239395.2
|
novel protein |
| chr1_-_247841896 | 0.18 |
ENST00000355784.3
|
OR11L1
|
olfactory receptor family 11 subfamily L member 1 |
| chr11_-_58228918 | 0.17 |
ENST00000316770.2
|
OR10Q1
|
olfactory receptor family 10 subfamily Q member 1 |
| chr2_-_27212522 | 0.17 |
ENST00000428518.5
|
SLC5A6
|
solute carrier family 5 member 6 |
| chr1_-_17045219 | 0.17 |
ENST00000491274.5
|
SDHB
|
succinate dehydrogenase complex iron sulfur subunit B |
| chrX_-_139205006 | 0.17 |
ENST00000436198.5
ENST00000626909.2 |
FGF13
|
fibroblast growth factor 13 |
| chr17_-_39927549 | 0.17 |
ENST00000579695.5
ENST00000304046.7 |
ORMDL3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr4_-_88823165 | 0.16 |
ENST00000508369.5
|
FAM13A
|
family with sequence similarity 13 member A |
| chr11_+_4994851 | 0.16 |
ENST00000641819.1
|
OR51L1
|
olfactory receptor family 51 subfamily L member 1 |
| chr21_+_43789522 | 0.15 |
ENST00000497547.2
|
RRP1
|
ribosomal RNA processing 1 |
| chr19_-_12734653 | 0.15 |
ENST00000592273.5
ENST00000588213.1 ENST00000242784.5 |
TRIR
|
telomerase RNA component interacting RNase |
| chr10_+_72893572 | 0.15 |
ENST00000622652.1
|
OIT3
|
oncoprotein induced transcript 3 |
| chr1_+_65147514 | 0.14 |
ENST00000545314.5
|
AK4
|
adenylate kinase 4 |
| chr5_+_55024250 | 0.14 |
ENST00000231009.3
|
GZMK
|
granzyme K |
| chr19_+_110643 | 0.13 |
ENST00000318050.4
|
OR4F17
|
olfactory receptor family 4 subfamily F member 17 |
| chr2_+_66435558 | 0.12 |
ENST00000488550.5
|
MEIS1
|
Meis homeobox 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FUBP1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 1.7 | 5.1 | GO:0044007 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 1.3 | 5.3 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.9 | 5.6 | GO:0001757 | somite specification(GO:0001757) |
| 0.7 | 9.5 | GO:0072564 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.7 | 4.8 | GO:0072011 | glomerular endothelium development(GO:0072011) |
| 0.6 | 2.4 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.6 | 1.8 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.5 | 2.6 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.5 | 9.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.4 | 2.2 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.4 | 8.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 4.6 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.4 | 2.5 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.4 | 2.8 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.4 | 1.2 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.4 | 1.6 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.4 | 4.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.4 | 1.8 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.4 | 4.0 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.3 | 12.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.3 | 1.0 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.3 | 2.0 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.3 | 1.1 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.3 | 1.4 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.3 | 1.8 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.3 | 1.5 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.2 | 2.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.2 | 2.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 6.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 1.0 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 1.0 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.2 | 3.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 1.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 1.0 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 3.4 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 3.4 | GO:0030903 | notochord development(GO:0030903) lateral mesoderm development(GO:0048368) |
| 0.1 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.3 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.7 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.9 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.5 | GO:0043375 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 | 1.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 3.0 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 3.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.1 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 3.1 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.1 | 1.0 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.3 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 0.4 | GO:0070378 | positive regulation of ERK5 cascade(GO:0070378) |
| 0.1 | 0.8 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 1.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.4 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.8 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.1 | 0.8 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 4.3 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.1 | 3.3 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.1 | 1.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.7 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 4.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.3 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.7 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 1.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.8 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 2.2 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.3 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 1.8 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 1.2 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.2 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.0 | 0.2 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.7 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 1.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.4 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 1.1 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 3.0 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.7 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 1.4 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 2.0 | GO:0065002 | intracellular protein transmembrane transport(GO:0065002) |
| 0.0 | 3.9 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.5 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.3 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.5 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 1.8 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 2.2 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.3 | GO:0035886 | vascular smooth muscle cell differentiation(GO:0035886) |
| 0.0 | 0.2 | GO:0046549 | photoreceptor cell outer segment organization(GO:0035845) retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.7 | 3.4 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.4 | 3.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 4.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.3 | 2.0 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.3 | 2.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.3 | 14.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 5.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.3 | 1.0 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.2 | 4.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 1.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 1.4 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 1.4 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.9 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 4.0 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 5.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 3.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.3 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 1.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 2.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 1.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.3 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 2.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.7 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 3.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.8 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 5.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 2.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 3.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.7 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 2.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 5.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 2.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.8 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 4.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 4.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 3.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.8 | 9.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.7 | 2.8 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.7 | 4.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.6 | 5.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.4 | 2.4 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.4 | 1.8 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 4.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 12.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 1.6 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 1.0 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.2 | 2.0 | GO:0034711 | inhibin binding(GO:0034711) BMP receptor activity(GO:0098821) |
| 0.2 | 1.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 1.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 3.4 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.2 | 4.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 1.0 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 0.5 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.2 | 1.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 1.4 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 2.5 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 1.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 1.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 1.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 3.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.7 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 2.5 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 2.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 1.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 2.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.3 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.1 | 4.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.4 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.3 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 13.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.7 | GO:0042835 | BRE binding(GO:0042835) |
| 0.1 | 1.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.4 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.1 | 1.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.1 | 3.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 2.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.5 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 1.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 4.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 4.3 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.7 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.9 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 5.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 1.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 1.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 4.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 3.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 2.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 10.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 7.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 3.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 4.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 4.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 5.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 2.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 6.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 8.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 4.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 2.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 12.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 9.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 4.8 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 2.0 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 5.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 2.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 9.7 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 5.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 7.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 3.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 7.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 3.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 2.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 5.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 3.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 2.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |