Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for GATA5
Z-value: 0.98
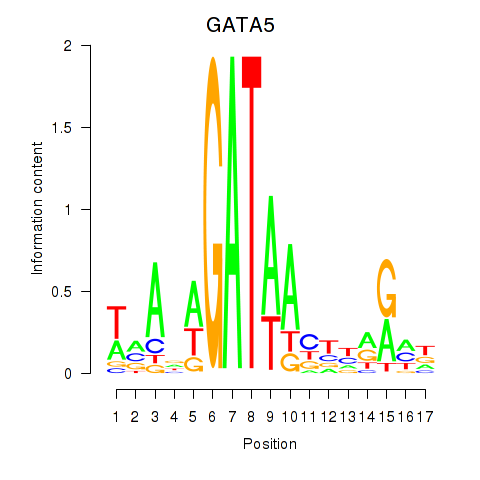
Transcription factors associated with GATA5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA5
|
ENSG00000130700.7 | GATA binding protein 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA5 | hg38_v1_chr20_-_62475983_62476001 | -0.31 | 8.6e-02 | Click! |
Activity profile of GATA5 motif
Sorted Z-values of GATA5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_71784046 | 5.41 |
ENST00000513476.5
ENST00000273951.13 |
GC
|
GC vitamin D binding protein |
| chr4_+_154563003 | 5.03 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr1_+_56854764 | 3.44 |
ENST00000361249.4
|
C8A
|
complement C8 alpha chain |
| chr2_+_101998955 | 2.56 |
ENST00000393414.6
|
IL1R2
|
interleukin 1 receptor type 2 |
| chr5_+_76609091 | 2.30 |
ENST00000514001.5
ENST00000396234.7 ENST00000509074.5 ENST00000502745.5 |
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr8_-_6999198 | 2.06 |
ENST00000382689.8
|
DEFA1B
|
defensin alpha 1B |
| chr17_+_7035979 | 2.01 |
ENST00000308027.7
|
SLC16A13
|
solute carrier family 16 member 13 |
| chr21_+_38296556 | 1.71 |
ENST00000398927.1
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr10_+_7703340 | 1.67 |
ENST00000429820.5
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr1_+_207104226 | 1.63 |
ENST00000367070.8
|
C4BPA
|
complement component 4 binding protein alpha |
| chr10_+_7703300 | 1.62 |
ENST00000358415.9
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr8_-_7018295 | 1.53 |
ENST00000327857.7
|
DEFA3
|
defensin alpha 3 |
| chr5_+_145936554 | 1.50 |
ENST00000359120.9
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr6_+_54099538 | 1.49 |
ENST00000447836.6
|
MLIP
|
muscular LMNA interacting protein |
| chr15_-_53733103 | 1.46 |
ENST00000559418.5
|
WDR72
|
WD repeat domain 72 |
| chr6_+_54099565 | 1.38 |
ENST00000511678.5
|
MLIP
|
muscular LMNA interacting protein |
| chr4_-_64409444 | 1.37 |
ENST00000381210.8
ENST00000507440.5 |
TECRL
|
trans-2,3-enoyl-CoA reductase like |
| chr8_-_65838730 | 1.36 |
ENST00000523253.1
|
PDE7A
|
phosphodiesterase 7A |
| chr12_+_20815672 | 1.29 |
ENST00000261196.6
ENST00000381541.7 ENST00000540229.1 |
SLCO1B3
SLCO1B3-SLCO1B7
|
solute carrier organic anion transporter family member 1B3 SLCO1B3-SLCO1B7 readthrough |
| chr15_+_58138368 | 1.26 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chr4_+_68815991 | 1.25 |
ENST00000265403.12
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase family 2 member B10 |
| chr4_-_64409381 | 1.24 |
ENST00000509536.1
|
TECRL
|
trans-2,3-enoyl-CoA reductase like |
| chr1_+_207104287 | 1.23 |
ENST00000421786.5
|
C4BPA
|
complement component 4 binding protein alpha |
| chrM_+_10759 | 1.23 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 4 |
| chrM_+_3298 | 1.19 |
ENST00000361390.2
|
MT-ND1
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 1 |
| chr6_-_132757883 | 1.19 |
ENST00000525289.5
ENST00000326499.11 |
VNN2
|
vanin 2 |
| chr6_-_132734692 | 1.08 |
ENST00000509351.5
ENST00000417437.6 ENST00000423615.6 ENST00000427187.6 ENST00000414302.7 ENST00000367927.9 ENST00000450865.2 |
VNN3
|
vanin 3 |
| chr1_+_174875505 | 0.99 |
ENST00000486220.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr6_-_134950081 | 0.99 |
ENST00000367847.2
ENST00000265605.7 ENST00000367845.6 |
ALDH8A1
|
aldehyde dehydrogenase 8 family member A1 |
| chrM_+_10464 | 0.99 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 4L |
| chr2_-_108989206 | 0.95 |
ENST00000258443.7
ENST00000409271.5 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr1_+_21570303 | 0.94 |
ENST00000374830.2
|
ALPL
|
alkaline phosphatase, biomineralization associated |
| chr15_+_76336755 | 0.92 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chr14_+_22007503 | 0.90 |
ENST00000390447.3
|
TRAV19
|
T cell receptor alpha variable 19 |
| chr7_+_142563730 | 0.89 |
ENST00000617347.1
|
TRBV12-4
|
T cell receptor beta variable 12-4 |
| chr7_+_142670734 | 0.82 |
ENST00000390398.3
|
TRBV25-1
|
T cell receptor beta variable 25-1 |
| chr9_-_35650902 | 0.82 |
ENST00000259608.8
ENST00000618781.1 |
SIT1
|
signaling threshold regulating transmembrane adaptor 1 |
| chr4_+_109912877 | 0.82 |
ENST00000265171.10
ENST00000509793.5 ENST00000652245.1 |
EGF
|
epidermal growth factor |
| chr19_+_55600277 | 0.81 |
ENST00000301073.4
|
ZNF524
|
zinc finger protein 524 |
| chr16_+_82034978 | 0.81 |
ENST00000563491.5
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2 |
| chr14_+_21768482 | 0.78 |
ENST00000390428.3
|
TRAV6
|
T cell receptor alpha variable 6 |
| chr7_+_142529268 | 0.78 |
ENST00000612787.1
|
TRBV7-9
|
T cell receptor beta variable 7-9 |
| chr14_+_19780242 | 0.77 |
ENST00000315957.4
|
OR4M1
|
olfactory receptor family 4 subfamily M member 1 |
| chr9_-_38720428 | 0.77 |
ENST00000637493.1
|
FAM240B
|
family with sequence similarity 240 member B |
| chr11_+_128693887 | 0.75 |
ENST00000281428.12
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr8_-_120812848 | 0.75 |
ENST00000520717.1
|
SNTB1
|
syntrophin beta 1 |
| chrM_+_12329 | 0.75 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 5 |
| chr14_+_35121814 | 0.75 |
ENST00000603611.5
|
PRORP
|
protein only RNase P catalytic subunit |
| chr11_+_112086853 | 0.74 |
ENST00000528182.5
ENST00000528048.5 ENST00000375549.8 ENST00000528021.6 ENST00000526592.5 ENST00000525291.5 |
SDHD
|
succinate dehydrogenase complex subunit D |
| chr12_+_8950036 | 0.73 |
ENST00000539240.5
|
KLRG1
|
killer cell lectin like receptor G1 |
| chr1_+_115029823 | 0.73 |
ENST00000256592.3
|
TSHB
|
thyroid stimulating hormone subunit beta |
| chr11_+_22672577 | 0.73 |
ENST00000534801.5
|
GAS2
|
growth arrest specific 2 |
| chr11_-_58440173 | 0.72 |
ENST00000302572.2
|
OR5B12
|
olfactory receptor family 5 subfamily B member 12 |
| chr8_+_40153475 | 0.70 |
ENST00000315792.5
|
TCIM
|
transcriptional and immune response regulator |
| chr11_-_63144221 | 0.70 |
ENST00000417740.5
ENST00000612278.4 ENST00000326192.5 |
SLC22A24
|
solute carrier family 22 member 24 |
| chrX_+_80420466 | 0.70 |
ENST00000308293.5
|
TENT5D
|
terminal nucleotidyltransferase 5D |
| chr3_-_126197932 | 0.68 |
ENST00000509952.5
|
ALDH1L1
|
aldehyde dehydrogenase 1 family member L1 |
| chr15_-_55408245 | 0.68 |
ENST00000563171.5
ENST00000425574.7 ENST00000442196.8 ENST00000564092.1 |
CCPG1
|
cell cycle progression 1 |
| chr3_-_197573323 | 0.68 |
ENST00000358186.6
ENST00000431056.5 |
BDH1
|
3-hydroxybutyrate dehydrogenase 1 |
| chr5_+_122129597 | 0.67 |
ENST00000514925.1
|
ENSG00000250803.6
|
novel zinc finger protein |
| chr3_-_108757407 | 0.66 |
ENST00000295755.7
|
RETNLB
|
resistin like beta |
| chr22_+_20859483 | 0.65 |
ENST00000439214.1
|
SNAP29
|
synaptosome associated protein 29 |
| chr3_+_172039556 | 0.65 |
ENST00000415807.7
ENST00000421757.5 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr11_+_55819630 | 0.64 |
ENST00000333976.7
|
OR5D18
|
olfactory receptor family 5 subfamily D member 18 |
| chr13_+_31739542 | 0.63 |
ENST00000380314.2
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr11_-_5986985 | 0.63 |
ENST00000332249.4
|
OR52L1
|
olfactory receptor family 52 subfamily L member 1 |
| chr3_+_46242453 | 0.63 |
ENST00000452454.1
ENST00000395940.3 ENST00000457243.1 |
CCR3
|
C-C motif chemokine receptor 3 |
| chr14_+_21924033 | 0.63 |
ENST00000390440.2
|
TRAV14DV4
|
T cell receptor alpha variable 14/delta variable 4 |
| chr5_-_59356962 | 0.63 |
ENST00000405755.6
|
PDE4D
|
phosphodiesterase 4D |
| chr14_-_21634940 | 0.62 |
ENST00000542433.1
|
OR10G2
|
olfactory receptor family 10 subfamily G member 2 |
| chr3_-_138132381 | 0.62 |
ENST00000236709.4
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr3_+_190615308 | 0.62 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr4_-_99657820 | 0.61 |
ENST00000511828.2
|
C4orf54
|
chromosome 4 open reading frame 54 |
| chr2_+_201132958 | 0.60 |
ENST00000479953.6
ENST00000340870.6 |
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr12_-_14814160 | 0.60 |
ENST00000316048.2
|
SMCO3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr12_-_110445540 | 0.60 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex subunit 3 |
| chr13_+_31739520 | 0.59 |
ENST00000298386.7
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr1_+_196774813 | 0.58 |
ENST00000471440.6
ENST00000391985.7 ENST00000617219.1 ENST00000367425.9 |
CFHR3
|
complement factor H related 3 |
| chr4_-_184773718 | 0.57 |
ENST00000505492.1
|
ACSL1
|
acyl-CoA synthetase long chain family member 1 |
| chr14_+_22052503 | 0.56 |
ENST00000390449.3
|
TRAV21
|
T cell receptor alpha variable 21 |
| chr5_-_154938181 | 0.56 |
ENST00000285873.8
|
GEMIN5
|
gem nuclear organelle associated protein 5 |
| chrX_-_154832671 | 0.56 |
ENST00000369529.1
|
SMIM9
|
small integral membrane protein 9 |
| chr11_+_56274154 | 0.55 |
ENST00000641665.1
|
OR5T1
|
olfactory receptor family 5 subfamily T member 1 |
| chr12_+_40310431 | 0.54 |
ENST00000681696.1
|
LRRK2
|
leucine rich repeat kinase 2 |
| chr9_-_6645712 | 0.54 |
ENST00000321612.8
|
GLDC
|
glycine decarboxylase |
| chr9_+_122742303 | 0.53 |
ENST00000304720.3
|
OR1L6
|
olfactory receptor family 1 subfamily L member 6 |
| chr20_+_45629717 | 0.53 |
ENST00000372643.4
|
WFDC10A
|
WAP four-disulfide core domain 10A |
| chr2_+_201132928 | 0.52 |
ENST00000462763.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr6_-_24935942 | 0.52 |
ENST00000645100.1
ENST00000643898.2 ENST00000613507.4 |
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr2_+_102311502 | 0.51 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr4_+_70637662 | 0.51 |
ENST00000472597.1
ENST00000472903.5 |
ENAM
ENSG00000286848.1
|
enamelin novel transcript |
| chr16_+_82035267 | 0.51 |
ENST00000566213.1
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2 |
| chr21_-_30480364 | 0.51 |
ENST00000390689.3
|
KRTAP19-1
|
keratin associated protein 19-1 |
| chr11_+_55723776 | 0.50 |
ENST00000641440.1
|
OR5D3P
|
olfactory receptor family 5 subfamily D member 3 pseudogene |
| chr17_+_22523404 | 0.50 |
ENST00000540040.3
|
MTRNR2L1
|
MT-RNR2 like 1 |
| chr8_+_85987287 | 0.50 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase H+ transporting V0 subunit d2 |
| chr6_+_106541111 | 0.50 |
ENST00000457437.1
|
CRYBG1
|
crystallin beta-gamma domain containing 1 |
| chrM_+_10055 | 0.49 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 3 |
| chr4_+_70195719 | 0.49 |
ENST00000683306.1
|
ODAM
|
odontogenic, ameloblast associated |
| chr7_+_142626642 | 0.48 |
ENST00000390394.3
|
TRBV20-1
|
T cell receptor beta variable 20-1 |
| chrX_-_77895546 | 0.48 |
ENST00000358075.11
|
MAGT1
|
magnesium transporter 1 |
| chr6_-_159044980 | 0.48 |
ENST00000367066.8
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr11_+_108224059 | 0.47 |
ENST00000601453.2
|
ATM
|
ATM serine/threonine kinase |
| chr4_-_109801978 | 0.47 |
ENST00000510800.1
ENST00000512148.5 ENST00000394634.7 ENST00000394635.8 ENST00000645635.1 |
CFI
ENSG00000285330.1
|
complement factor I novel protein |
| chr8_-_85341659 | 0.47 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr11_+_5893208 | 0.47 |
ENST00000610445.2
|
OR52E5
|
olfactory receptor family 52 subfamily E member 5 |
| chr9_+_72628020 | 0.46 |
ENST00000646619.1
|
TMC1
|
transmembrane channel like 1 |
| chr11_+_56103687 | 0.46 |
ENST00000313503.2
ENST00000641311.1 |
OR8H2
|
olfactory receptor family 8 subfamily H member 2 |
| chr3_+_46242371 | 0.46 |
ENST00000545097.1
|
CCR3
|
C-C motif chemokine receptor 3 |
| chr6_+_29396555 | 0.46 |
ENST00000623183.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2 |
| chr17_+_38752731 | 0.45 |
ENST00000619426.5
ENST00000610434.4 |
PSMB3
|
proteasome 20S subunit beta 3 |
| chr11_+_124183219 | 0.45 |
ENST00000641351.2
|
OR10D3
|
olfactory receptor family 10 subfamily D member 3 |
| chr4_+_145625992 | 0.45 |
ENST00000541599.5
|
MMAA
|
metabolism of cobalamin associated A |
| chr8_+_7495892 | 0.45 |
ENST00000355602.3
|
DEFB107B
|
defensin beta 107B |
| chr19_+_926001 | 0.45 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr3_+_108822778 | 0.44 |
ENST00000295756.11
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr4_-_82844418 | 0.44 |
ENST00000503937.5
|
SEC31A
|
SEC31 homolog A, COPII coat complex component |
| chr6_+_18387326 | 0.44 |
ENST00000259939.4
|
RNF144B
|
ring finger protein 144B |
| chr1_+_86468902 | 0.44 |
ENST00000394711.2
|
CLCA1
|
chloride channel accessory 1 |
| chr1_-_159536007 | 0.44 |
ENST00000334857.3
|
OR10J5
|
olfactory receptor family 10 subfamily J member 5 |
| chrX_-_33339525 | 0.44 |
ENST00000288447.9
|
DMD
|
dystrophin |
| chr11_+_4593750 | 0.44 |
ENST00000450052.3
|
OR52I1
|
olfactory receptor family 52 subfamily I member 1 |
| chr10_-_61001430 | 0.44 |
ENST00000357917.4
|
RHOBTB1
|
Rho related BTB domain containing 1 |
| chr12_-_68225806 | 0.43 |
ENST00000229134.5
|
IL26
|
interleukin 26 |
| chr4_+_70592253 | 0.43 |
ENST00000322937.10
ENST00000613447.4 |
AMBN
|
ameloblastin |
| chr15_+_21637909 | 0.43 |
ENST00000332663.3
|
OR4M2B
|
olfactory receptor family 4 subfamily M member 2B |
| chr7_+_18290416 | 0.43 |
ENST00000413509.6
|
HDAC9
|
histone deacetylase 9 |
| chr9_+_122723990 | 0.43 |
ENST00000259466.1
|
OR1L4
|
olfactory receptor family 1 subfamily L member 4 |
| chr15_-_49620929 | 0.42 |
ENST00000561064.5
|
FAM227B
|
family with sequence similarity 227 member B |
| chr16_-_68015911 | 0.42 |
ENST00000574912.1
|
DPEP2NB
|
DPEP2 neighbor |
| chr4_-_99435134 | 0.41 |
ENST00000476959.5
ENST00000482593.5 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr14_+_21317535 | 0.41 |
ENST00000382933.8
|
RPGRIP1
|
RPGR interacting protein 1 |
| chr3_+_108822759 | 0.41 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr16_+_31527876 | 0.41 |
ENST00000302312.9
ENST00000564707.1 |
AHSP
|
alpha hemoglobin stabilizing protein |
| chr12_+_57488091 | 0.40 |
ENST00000630571.2
ENST00000630803.1 |
MARS1
|
methionyl-tRNA synthetase 1 |
| chr4_-_68129853 | 0.40 |
ENST00000356291.3
|
TMPRSS11F
|
transmembrane serine protease 11F |
| chr5_+_148063971 | 0.40 |
ENST00000398454.5
ENST00000359874.7 ENST00000508733.5 ENST00000256084.8 |
SPINK5
|
serine peptidase inhibitor Kazal type 5 |
| chr1_-_158686700 | 0.39 |
ENST00000643759.2
|
SPTA1
|
spectrin alpha, erythrocytic 1 |
| chr8_+_27771942 | 0.39 |
ENST00000523566.5
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr7_+_142313144 | 0.38 |
ENST00000390357.3
|
TRBV4-1
|
T cell receptor beta variable 4-1 |
| chr5_+_75653161 | 0.37 |
ENST00000672390.1
|
ANKDD1B
|
ankyrin repeat and death domain containing 1B |
| chr2_-_49154433 | 0.37 |
ENST00000454032.5
ENST00000304421.8 |
FSHR
|
follicle stimulating hormone receptor |
| chr1_+_212301806 | 0.37 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2 regulatory subunit B'alpha |
| chr4_-_47981535 | 0.37 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr11_-_101129706 | 0.36 |
ENST00000534013.5
|
PGR
|
progesterone receptor |
| chr17_-_76643648 | 0.36 |
ENST00000589992.1
|
ST6GALNAC1
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 |
| chr2_-_135837170 | 0.36 |
ENST00000264162.7
|
LCT
|
lactase |
| chr2_+_201132872 | 0.36 |
ENST00000470178.6
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr14_-_24084625 | 0.36 |
ENST00000397002.6
|
NRL
|
neural retina leucine zipper |
| chrX_-_77895414 | 0.36 |
ENST00000618282.5
ENST00000373336.3 |
MAGT1
|
magnesium transporter 1 |
| chr11_-_4924339 | 0.35 |
ENST00000623849.1
|
OR51G1
|
olfactory receptor family 51 subfamily G member 1 |
| chr10_+_5048748 | 0.35 |
ENST00000602997.5
ENST00000439082.7 |
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr11_+_30231000 | 0.35 |
ENST00000254122.8
ENST00000533718.2 ENST00000417547.1 |
FSHB
|
follicle stimulating hormone subunit beta |
| chr15_+_85689869 | 0.35 |
ENST00000557852.2
|
AKAP13
|
A-kinase anchoring protein 13 |
| chr2_+_102311546 | 0.35 |
ENST00000233954.6
ENST00000447231.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr4_-_159035226 | 0.35 |
ENST00000434826.3
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr18_+_63775395 | 0.35 |
ENST00000398019.7
|
SERPINB7
|
serpin family B member 7 |
| chr4_+_174283699 | 0.35 |
ENST00000505124.5
|
CEP44
|
centrosomal protein 44 |
| chr17_-_47831509 | 0.34 |
ENST00000414011.1
ENST00000351111.7 |
MRPL10
|
mitochondrial ribosomal protein L10 |
| chr7_+_142791635 | 0.33 |
ENST00000633705.1
|
TRBC1
|
T cell receptor beta constant 1 |
| chr11_+_124241095 | 0.33 |
ENST00000641972.1
|
OR8G1
|
olfactory receptor family 8 subfamily G member 1 |
| chr15_-_34336749 | 0.32 |
ENST00000397707.6
ENST00000560611.5 |
SLC12A6
|
solute carrier family 12 member 6 |
| chr19_-_19628512 | 0.32 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr17_-_4786354 | 0.32 |
ENST00000328739.6
ENST00000441199.2 ENST00000416307.6 |
VMO1
|
vitelline membrane outer layer 1 homolog |
| chr7_+_144095048 | 0.32 |
ENST00000408949.2
|
OR2A12
|
olfactory receptor family 2 subfamily A member 12 |
| chr6_+_132552693 | 0.32 |
ENST00000275200.1
|
TAAR8
|
trace amine associated receptor 8 |
| chr6_+_29461440 | 0.31 |
ENST00000396792.2
|
OR2H1
|
olfactory receptor family 2 subfamily H member 1 |
| chr20_-_45579277 | 0.31 |
ENST00000289953.3
|
WFDC8
|
WAP four-disulfide core domain 8 |
| chr1_+_42787990 | 0.31 |
ENST00000536543.5
|
TMEM269
|
transmembrane protein 269 |
| chr4_+_87608529 | 0.31 |
ENST00000651931.1
|
DSPP
|
dentin sialophosphoprotein |
| chr7_+_18290089 | 0.31 |
ENST00000433709.6
|
HDAC9
|
histone deacetylase 9 |
| chr2_-_49154507 | 0.30 |
ENST00000406846.7
|
FSHR
|
follicle stimulating hormone receptor |
| chr10_+_15043919 | 0.30 |
ENST00000378225.5
|
OLAH
|
oleoyl-ACP hydrolase |
| chr15_-_49620747 | 0.30 |
ENST00000559905.5
ENST00000560246.1 ENST00000299338.11 ENST00000558594.5 |
FAM227B
|
family with sequence similarity 227 member B |
| chr4_-_99435336 | 0.30 |
ENST00000437033.7
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr11_+_122862303 | 0.30 |
ENST00000533709.1
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr1_+_23743153 | 0.29 |
ENST00000418390.6
|
ELOA
|
elongin A |
| chr17_-_4786433 | 0.29 |
ENST00000354194.4
|
VMO1
|
vitelline membrane outer layer 1 homolog |
| chr13_-_102759059 | 0.29 |
ENST00000322527.4
|
CCDC168
|
coiled-coil domain containing 168 |
| chr13_+_108218366 | 0.28 |
ENST00000375898.4
|
ABHD13
|
abhydrolase domain containing 13 |
| chrX_+_27590400 | 0.28 |
ENST00000431122.2
|
DCAF8L2
|
DDB1 and CUL4 associated factor 8 like 2 |
| chr6_+_146029131 | 0.28 |
ENST00000355289.8
|
GRM1
|
glutamate metabotropic receptor 1 |
| chrX_+_1615158 | 0.28 |
ENST00000381229.9
ENST00000381233.8 |
ASMT
|
acetylserotonin O-methyltransferase |
| chr17_-_41047267 | 0.27 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr1_-_44141631 | 0.27 |
ENST00000634670.1
|
KLF18
|
Kruppel like factor 18 |
| chr19_-_41844629 | 0.27 |
ENST00000609812.6
|
LYPD4
|
LY6/PLAUR domain containing 4 |
| chr1_-_247716646 | 0.27 |
ENST00000641444.1
ENST00000641470.1 |
OR6F1
|
olfactory receptor family 6 subfamily F member 1 |
| chr1_+_248375746 | 0.26 |
ENST00000641644.1
|
OR2T6
|
olfactory receptor family 2 subfamily T member 6 |
| chr2_+_178284907 | 0.26 |
ENST00000409631.5
|
OSBPL6
|
oxysterol binding protein like 6 |
| chrX_+_153179276 | 0.26 |
ENST00000356661.7
|
MAGEA1
|
MAGE family member A1 |
| chr19_-_9214871 | 0.26 |
ENST00000308682.3
|
OR7D4
|
olfactory receptor family 7 subfamily D member 4 |
| chr12_+_71686473 | 0.26 |
ENST00000549735.5
|
TMEM19
|
transmembrane protein 19 |
| chr2_+_96642729 | 0.26 |
ENST00000624922.6
ENST00000623019.5 |
FER1L5
|
fer-1 like family member 5 |
| chr17_+_43170452 | 0.25 |
ENST00000341165.10
ENST00000586650.5 |
NBR1
|
NBR1 autophagy cargo receptor |
| chr14_+_19875142 | 0.25 |
ENST00000641885.1
|
OR4K2
|
olfactory receptor family 4 subfamily K member 2 |
| chr1_+_248097276 | 0.25 |
ENST00000641714.1
|
OR2L13
|
olfactory receptor family 2 subfamily L member 13 |
| chr3_-_146528750 | 0.25 |
ENST00000483300.5
|
PLSCR1
|
phospholipid scramblase 1 |
| chr2_-_85663393 | 0.24 |
ENST00000494165.1
|
SFTPB
|
surfactant protein B |
| chr15_+_40952962 | 0.24 |
ENST00000444189.7
|
CHAC1
|
ChaC glutathione specific gamma-glutamylcyclotransferase 1 |
| chr8_+_12108172 | 0.24 |
ENST00000400078.3
|
ZNF705D
|
zinc finger protein 705D |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.4 | 5.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 2.9 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.3 | 0.9 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.3 | 0.9 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.3 | 1.3 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.2 | 0.9 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 3.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 1.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 0.7 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.2 | 0.5 | GO:1903123 | regulation of thioredoxin peroxidase activity(GO:1903123) negative regulation of thioredoxin peroxidase activity(GO:1903124) negative regulation of thioredoxin peroxidase activity by peptidyl-threonine phosphorylation(GO:1903125) Wnt signalosome assembly(GO:1904887) negative regulation of peroxidase activity(GO:2000469) |
| 0.2 | 0.5 | GO:0036255 | response to methylamine(GO:0036255) response to lipoic acid(GO:1903442) |
| 0.2 | 0.4 | GO:0046137 | negative regulation of vitamin metabolic process(GO:0046137) |
| 0.2 | 0.5 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.2 | 0.7 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.2 | 0.7 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.2 | 5.4 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 0.8 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.1 | 2.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.4 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.7 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 1.6 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 1.0 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.9 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 2.6 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 1.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 1.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.5 | GO:1904882 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.4 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 1.5 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 2.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.4 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.2 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.1 | 1.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 4.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.9 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.4 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.6 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.4 | GO:0002786 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.1 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.7 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:1902990 | mitochondrial DNA repair(GO:0043504) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.5 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.3 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.5 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.2 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 2.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.0 | 0.4 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 1.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.2 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.2 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.4 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 9.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:2000870 | oocyte growth(GO:0001555) regulation of progesterone secretion(GO:2000870) |
| 0.0 | 1.2 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.7 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.3 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0070510 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 1.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.5 | GO:0090662 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.3 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0002760 | positive regulation of antimicrobial humoral response(GO:0002760) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 3.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 0.5 | GO:0032473 | cytoplasmic side of mitochondrial outer membrane(GO:0032473) caveola neck(GO:0099400) |
| 0.1 | 0.7 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.5 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.1 | 0.5 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.9 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.7 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 0.2 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.1 | 0.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 1.6 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 4.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.6 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 12.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.6 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 2.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.2 | GO:0097208 | alveolar lamellar body(GO:0097208) multivesicular body lumen(GO:0097486) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.7 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 2.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.4 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.6 | 2.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.5 | 2.6 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.3 | 0.9 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.3 | 1.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.3 | 1.3 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.2 | 0.9 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.2 | 0.7 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.2 | 0.5 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) peroxidase inhibitor activity(GO:0036479) |
| 0.2 | 0.5 | GO:0004375 | glycine dehydrogenase (decarboxylating) activity(GO:0004375) oxidoreductase activity, acting on the CH-NH2 group of donors, disulfide as acceptor(GO:0016642) vitamin B6 binding(GO:0070279) pyridoxal binding(GO:0070280) |
| 0.2 | 0.7 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.2 | 0.7 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.2 | 0.7 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 2.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.9 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.6 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 0.4 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.1 | 0.6 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.1 | 0.9 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 3.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.4 | GO:0016296 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.1 | 4.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.3 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.5 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.5 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 1.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.6 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 2.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.8 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 2.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 2.6 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 2.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 5.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 4.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 6.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 3.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 2.9 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 3.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 3.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 5.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 4.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 1.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 3.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 7.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.9 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 3.2 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |