Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
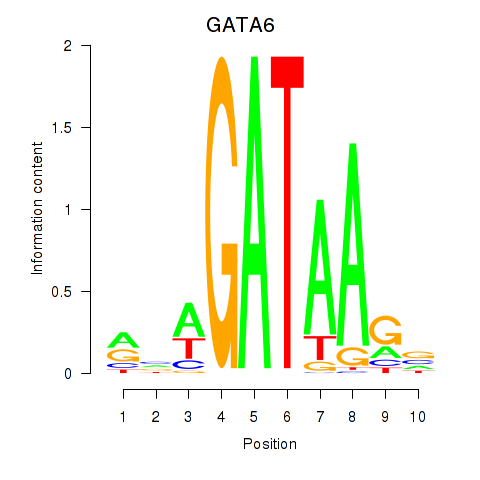
Results for GATA6
Z-value: 1.23
Transcription factors associated with GATA6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA6
|
ENSG00000141448.11 | GATA binding protein 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA6 | hg38_v1_chr18_+_22169580_22169596 | -0.02 | 9.3e-01 | Click! |
Activity profile of GATA6 motif
Sorted Z-values of GATA6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_102311502 | 5.85 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr10_-_97771954 | 5.19 |
ENST00000266066.4
|
SFRP5
|
secreted frizzled related protein 5 |
| chr13_-_46142834 | 4.56 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr2_+_102311546 | 4.15 |
ENST00000233954.6
ENST00000447231.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr12_-_54295748 | 3.82 |
ENST00000540264.2
ENST00000312156.8 |
NFE2
|
nuclear factor, erythroid 2 |
| chr11_-_116837586 | 3.67 |
ENST00000375320.5
ENST00000359492.6 ENST00000375329.6 ENST00000375323.5 ENST00000236850.5 |
APOA1
|
apolipoprotein A1 |
| chr7_-_100641507 | 3.66 |
ENST00000431692.5
ENST00000223051.8 |
TFR2
|
transferrin receptor 2 |
| chr9_-_114930508 | 3.51 |
ENST00000223795.3
ENST00000618336.4 |
TNFSF8
|
TNF superfamily member 8 |
| chr17_+_4771878 | 3.48 |
ENST00000270560.4
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr1_-_167518583 | 3.29 |
ENST00000392122.3
|
CD247
|
CD247 molecule |
| chr12_-_9999176 | 3.05 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr1_-_26913964 | 3.03 |
ENST00000254227.4
|
NR0B2
|
nuclear receptor subfamily 0 group B member 2 |
| chr1_-_167518521 | 2.95 |
ENST00000362089.10
|
CD247
|
CD247 molecule |
| chr19_+_7677082 | 2.87 |
ENST00000597445.1
|
MCEMP1
|
mast cell expressed membrane protein 1 |
| chr17_-_31318818 | 2.60 |
ENST00000578584.5
|
ENSG00000265118.5
|
novel protein |
| chr7_+_155298561 | 2.45 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr15_-_38560050 | 2.37 |
ENST00000558432.5
|
RASGRP1
|
RAS guanyl releasing protein 1 |
| chr10_-_127892930 | 2.14 |
ENST00000368671.4
|
CLRN3
|
clarin 3 |
| chr13_-_40982880 | 2.04 |
ENST00000635415.1
|
ELF1
|
E74 like ETS transcription factor 1 |
| chr1_-_225999312 | 1.89 |
ENST00000272091.8
|
SDE2
|
SDE2 telomere maintenance homolog |
| chr16_+_31527876 | 1.88 |
ENST00000302312.9
ENST00000564707.1 |
AHSP
|
alpha hemoglobin stabilizing protein |
| chr3_+_137998735 | 1.86 |
ENST00000343735.8
|
CLDN18
|
claudin 18 |
| chr19_+_10286971 | 1.81 |
ENST00000340992.4
ENST00000393717.2 |
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr17_-_35560812 | 1.73 |
ENST00000674182.1
|
SLFN14
|
schlafen family member 14 |
| chr14_-_22815421 | 1.70 |
ENST00000674313.1
ENST00000555959.1 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr11_+_75717811 | 1.70 |
ENST00000198801.10
|
MOGAT2
|
monoacylglycerol O-acyltransferase 2 |
| chr11_-_47352693 | 1.63 |
ENST00000256993.8
ENST00000399249.6 ENST00000545968.6 |
MYBPC3
|
myosin binding protein C3 |
| chr4_-_102345196 | 1.61 |
ENST00000683412.1
ENST00000682227.1 |
SLC39A8
|
solute carrier family 39 member 8 |
| chr6_+_31706866 | 1.58 |
ENST00000375832.5
ENST00000503322.1 |
LY6G6F
LY6G6F-LY6G6D
|
lymphocyte antigen 6 family member G6F LY6G6F-LY6G6D readthrough |
| chr19_+_10286944 | 1.55 |
ENST00000380770.5
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr19_-_38812936 | 1.54 |
ENST00000307751.9
ENST00000594209.1 |
LGALS4
|
galectin 4 |
| chr12_+_111405861 | 1.54 |
ENST00000341259.7
|
SH2B3
|
SH2B adaptor protein 3 |
| chr12_-_102950835 | 1.53 |
ENST00000546844.1
|
PAH
|
phenylalanine hydroxylase |
| chr4_-_102345061 | 1.47 |
ENST00000394833.6
|
SLC39A8
|
solute carrier family 39 member 8 |
| chr11_-_57390636 | 1.47 |
ENST00000525955.1
ENST00000533605.5 ENST00000311862.10 |
PRG2
|
proteoglycan 2, pro eosinophil major basic protein |
| chr4_+_99574812 | 1.47 |
ENST00000422897.6
ENST00000265517.10 |
MTTP
|
microsomal triglyceride transfer protein |
| chr14_+_105486311 | 1.46 |
ENST00000330233.11
|
CRIP1
|
cysteine rich protein 1 |
| chr9_-_112175185 | 1.44 |
ENST00000355396.7
|
SUSD1
|
sushi domain containing 1 |
| chr2_+_43838963 | 1.41 |
ENST00000272286.4
|
ABCG8
|
ATP binding cassette subfamily G member 8 |
| chr2_-_68871382 | 1.41 |
ENST00000295379.2
|
BMP10
|
bone morphogenetic protein 10 |
| chr22_-_19881369 | 1.40 |
ENST00000462330.5
|
TXNRD2
|
thioredoxin reductase 2 |
| chr17_+_4932248 | 1.40 |
ENST00000329125.6
|
GP1BA
|
glycoprotein Ib platelet subunit alpha |
| chr17_-_49764123 | 1.39 |
ENST00000240364.7
ENST00000506156.1 |
FAM117A
|
family with sequence similarity 117 member A |
| chr2_-_43838832 | 1.37 |
ENST00000405322.8
|
ABCG5
|
ATP binding cassette subfamily G member 5 |
| chr22_-_19881163 | 1.34 |
ENST00000485358.5
|
TXNRD2
|
thioredoxin reductase 2 |
| chr19_-_13102848 | 1.32 |
ENST00000264824.5
|
LYL1
|
LYL1 basic helix-loop-helix family member |
| chr14_+_75522427 | 1.32 |
ENST00000286639.8
|
BATF
|
basic leucine zipper ATF-like transcription factor |
| chr13_-_27969295 | 1.27 |
ENST00000381020.8
|
CDX2
|
caudal type homeobox 2 |
| chr9_-_112175264 | 1.22 |
ENST00000374264.6
|
SUSD1
|
sushi domain containing 1 |
| chr17_+_4932285 | 1.18 |
ENST00000611961.1
|
GP1BA
|
glycoprotein Ib platelet subunit alpha |
| chr14_+_75522531 | 1.16 |
ENST00000555504.1
|
BATF
|
basic leucine zipper ATF-like transcription factor |
| chr6_-_30160880 | 1.12 |
ENST00000376704.3
|
TRIM10
|
tripartite motif containing 10 |
| chr4_-_48681171 | 1.11 |
ENST00000505759.1
|
FRYL
|
FRY like transcription coactivator |
| chr9_-_112175283 | 1.11 |
ENST00000374270.8
ENST00000374263.7 |
SUSD1
|
sushi domain containing 1 |
| chr18_-_31162849 | 1.09 |
ENST00000257197.7
ENST00000257198.6 |
DSC1
|
desmocollin 1 |
| chr14_-_22815493 | 1.08 |
ENST00000555911.1
|
SLC7A7
|
solute carrier family 7 member 7 |
| chr17_+_77287891 | 1.08 |
ENST00000449803.6
ENST00000431235.6 |
SEPTIN9
|
septin 9 |
| chr14_-_22815856 | 1.02 |
ENST00000554758.1
ENST00000397528.8 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr5_-_135954962 | 0.98 |
ENST00000522943.5
ENST00000514447.2 ENST00000274507.6 |
LECT2
|
leukocyte cell derived chemotaxin 2 |
| chr11_+_10305370 | 0.97 |
ENST00000528544.1
|
ADM
|
adrenomedullin |
| chr3_+_148865288 | 0.96 |
ENST00000296046.4
|
CPA3
|
carboxypeptidase A3 |
| chr14_-_22815801 | 0.96 |
ENST00000397532.9
|
SLC7A7
|
solute carrier family 7 member 7 |
| chr18_-_2982873 | 0.95 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr9_-_97938157 | 0.87 |
ENST00000616898.2
|
HEMGN
|
hemogen |
| chr20_+_44401269 | 0.86 |
ENST00000443598.6
ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4 alpha |
| chr8_-_20183090 | 0.85 |
ENST00000265808.11
ENST00000522513.5 |
SLC18A1
|
solute carrier family 18 member A1 |
| chr17_+_76385256 | 0.85 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr2_-_40512423 | 0.85 |
ENST00000402441.5
ENST00000448531.1 |
SLC8A1
|
solute carrier family 8 member A1 |
| chr2_-_201433545 | 0.83 |
ENST00000440597.1
|
TRAK2
|
trafficking kinesin protein 2 |
| chr7_-_78770859 | 0.81 |
ENST00000636717.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr12_-_52601397 | 0.80 |
ENST00000549979.1
|
KRT72
|
keratin 72 |
| chr17_-_64006880 | 0.79 |
ENST00000449662.6
|
ICAM2
|
intercellular adhesion molecule 2 |
| chr18_+_22169580 | 0.78 |
ENST00000269216.10
|
GATA6
|
GATA binding protein 6 |
| chr6_-_30161200 | 0.78 |
ENST00000449742.7
|
TRIM10
|
tripartite motif containing 10 |
| chr8_-_20183127 | 0.77 |
ENST00000276373.10
ENST00000519026.5 ENST00000440926.3 ENST00000437980.3 |
SLC18A1
|
solute carrier family 18 member A1 |
| chr11_+_10305065 | 0.73 |
ENST00000534464.1
ENST00000278175.10 ENST00000530439.1 ENST00000524948.5 ENST00000528655.5 ENST00000526492.4 ENST00000525063.2 |
ADM
|
adrenomedullin |
| chr2_-_40512361 | 0.71 |
ENST00000403092.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr5_-_142324137 | 0.70 |
ENST00000511815.1
|
SPRY4
|
sprouty RTK signaling antagonist 4 |
| chr22_-_29388530 | 0.70 |
ENST00000357586.7
ENST00000432560.6 ENST00000405198.6 ENST00000317368.11 |
AP1B1
|
adaptor related protein complex 1 subunit beta 1 |
| chr6_+_117675448 | 0.69 |
ENST00000368494.4
|
NUS1
|
NUS1 dehydrodolichyl diphosphate synthase subunit |
| chr3_+_152269103 | 0.67 |
ENST00000459747.1
|
MBNL1
|
muscleblind like splicing regulator 1 |
| chr8_-_92095432 | 0.67 |
ENST00000518954.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr6_+_138377905 | 0.66 |
ENST00000573100.2
|
SMIM28
|
small integral membrane protein 28 |
| chr19_-_13103151 | 0.66 |
ENST00000590974.1
|
LYL1
|
LYL1 basic helix-loop-helix family member |
| chr19_-_11384299 | 0.63 |
ENST00000592375.6
ENST00000222139.11 |
EPOR
|
erythropoietin receptor |
| chr1_+_25272527 | 0.60 |
ENST00000342055.9
ENST00000357542.8 ENST00000417538.6 ENST00000423810.6 ENST00000568195.5 |
RHD
|
Rh blood group D antigen |
| chr2_+_168901290 | 0.59 |
ENST00000429379.2
ENST00000375363.8 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase catalytic subunit 2 |
| chrX_+_108045050 | 0.55 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr6_+_130018565 | 0.54 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr8_-_92095498 | 0.51 |
ENST00000520974.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr18_+_3447562 | 0.49 |
ENST00000618001.4
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr8_-_92095410 | 0.48 |
ENST00000518823.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr19_-_55038256 | 0.48 |
ENST00000417454.5
ENST00000310373.7 ENST00000333884.2 |
GP6
|
glycoprotein VI platelet |
| chr2_-_213150387 | 0.47 |
ENST00000433134.5
|
IKZF2
|
IKAROS family zinc finger 2 |
| chr17_+_19411118 | 0.46 |
ENST00000575165.6
|
RNF112
|
ring finger protein 112 |
| chr2_-_213150236 | 0.45 |
ENST00000442445.1
ENST00000342002.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr4_-_185775484 | 0.41 |
ENST00000444771.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr12_+_71938837 | 0.35 |
ENST00000333850.4
|
TPH2
|
tryptophan hydroxylase 2 |
| chr2_+_118096418 | 0.34 |
ENST00000614681.1
|
INSIG2
|
insulin induced gene 2 |
| chr4_+_165873231 | 0.34 |
ENST00000061240.7
|
TLL1
|
tolloid like 1 |
| chr20_-_32207708 | 0.32 |
ENST00000246229.5
|
PLAGL2
|
PLAG1 like zinc finger 2 |
| chr1_+_50108856 | 0.31 |
ENST00000650764.1
ENST00000494555.2 ENST00000371824.7 ENST00000371823.8 ENST00000652693.1 |
ELAVL4
|
ELAV like RNA binding protein 4 |
| chrX_+_38006551 | 0.31 |
ENST00000297875.7
|
SYTL5
|
synaptotagmin like 5 |
| chrX_+_108044967 | 0.29 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr4_-_185775890 | 0.25 |
ENST00000437304.6
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr10_+_116545907 | 0.25 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr19_+_14028148 | 0.25 |
ENST00000431365.3
ENST00000585987.1 |
RLN3
|
relaxin 3 |
| chr1_-_205943449 | 0.24 |
ENST00000367135.8
ENST00000367134.2 |
SLC26A9
|
solute carrier family 26 member 9 |
| chr2_+_137964446 | 0.24 |
ENST00000280096.5
ENST00000280097.5 |
HNMT
|
histamine N-methyltransferase |
| chr8_-_144792380 | 0.22 |
ENST00000532702.1
ENST00000394920.6 ENST00000528957.6 ENST00000527914.5 |
RPL8
|
ribosomal protein L8 |
| chr11_-_66438788 | 0.18 |
ENST00000329819.4
ENST00000310999.11 ENST00000430466.6 |
MRPL11
|
mitochondrial ribosomal protein L11 |
| chr3_+_152268920 | 0.18 |
ENST00000495875.6
ENST00000324210.10 ENST00000493459.5 |
MBNL1
|
muscleblind like splicing regulator 1 |
| chr14_-_21476874 | 0.17 |
ENST00000649801.1
ENST00000397762.6 |
RAB2B
|
RAB2B, member RAS oncogene family |
| chr1_+_154321076 | 0.16 |
ENST00000324978.8
|
AQP10
|
aquaporin 10 |
| chr2_+_172928165 | 0.16 |
ENST00000535187.5
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr4_+_165873348 | 0.15 |
ENST00000507499.5
|
TLL1
|
tolloid like 1 |
| chr1_-_206023889 | 0.13 |
ENST00000358184.7
ENST00000360218.3 ENST00000678712.1 ENST00000678498.1 |
CTSE
|
cathepsin E |
| chr8_-_92095468 | 0.13 |
ENST00000520583.5
ENST00000519061.5 |
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr3_-_122975369 | 0.12 |
ENST00000449546.1
|
SEMA5B
|
semaphorin 5B |
| chr6_-_154247630 | 0.11 |
ENST00000519344.5
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr8_+_69466837 | 0.09 |
ENST00000529134.5
|
SULF1
|
sulfatase 1 |
| chr20_+_45306834 | 0.08 |
ENST00000343694.8
ENST00000372741.7 ENST00000372743.5 |
RBPJL
|
recombination signal binding protein for immunoglobulin kappa J region like |
| chr5_+_1201588 | 0.08 |
ENST00000304460.11
|
SLC6A19
|
solute carrier family 6 member 19 |
| chr7_+_142749465 | 0.07 |
ENST00000486171.5
ENST00000619214.4 ENST00000311737.12 |
PRSS1
|
serine protease 1 |
| chr5_-_33984681 | 0.07 |
ENST00000296589.9
|
SLC45A2
|
solute carrier family 45 member 2 |
| chr16_+_1431075 | 0.05 |
ENST00000640283.2
|
PERCC1
|
proline and glutamate rich with coiled coil 1 |
| chr20_+_56392353 | 0.05 |
ENST00000415828.5
ENST00000428552.1 |
CSTF1
|
cleavage stimulation factor subunit 1 |
| chr10_+_116590956 | 0.04 |
ENST00000358834.9
ENST00000528052.5 |
PNLIPRP1
|
pancreatic lipase related protein 1 |
| chr7_+_150993773 | 0.04 |
ENST00000467517.1
ENST00000484524.5 |
NOS3
|
nitric oxide synthase 3 |
| chr1_+_174877430 | 0.04 |
ENST00000392064.6
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr10_+_116591010 | 0.02 |
ENST00000530319.5
ENST00000527980.5 ENST00000471549.5 ENST00000534537.5 |
PNLIPRP1
|
pancreatic lipase related protein 1 |
| chr19_+_35358821 | 0.01 |
ENST00000594310.1
|
FFAR3
|
free fatty acid receptor 3 |
| chr1_+_154321107 | 0.00 |
ENST00000484864.1
|
AQP10
|
aquaporin 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 10.0 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.8 | 2.4 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.7 | 5.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.7 | 3.7 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.7 | 2.8 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.6 | 3.7 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.5 | 4.8 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.4 | 1.5 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.4 | 1.5 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.3 | 2.4 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.3 | 0.8 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.3 | 3.1 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.3 | 1.6 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.2 | 1.7 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 1.6 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.2 | 0.7 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.2 | 0.6 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.2 | 1.8 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.2 | 0.8 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.2 | 0.4 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.2 | 4.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 1.9 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.2 | 1.7 | GO:0097647 | progesterone biosynthetic process(GO:0006701) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 3.5 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.8 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.1 | 1.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 6.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.6 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 1.4 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.1 | 1.7 | GO:0071287 | cellular response to manganese ion(GO:0071287) |
| 0.1 | 2.5 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 2.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 3.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 2.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 1.5 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.9 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.8 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.1 | 3.1 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 2.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.9 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 1.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 1.5 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 2.8 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 1.0 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.0 | 0.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 3.0 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.3 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.3 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.5 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.7 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 1.8 | GO:0070268 | cornification(GO:0070268) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.6 | 2.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.5 | 6.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.4 | 1.6 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.2 | 3.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 2.8 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 1.6 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.1 | 4.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 2.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 1.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 9.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 3.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 10.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 1.2 | 3.7 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.7 | 3.7 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.4 | 1.6 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.3 | 2.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 1.9 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 1.5 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.2 | 1.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 4.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 0.6 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.2 | 5.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 1.7 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.2 | 1.6 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.2 | 0.9 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 2.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.2 | 3.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.8 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 1.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 2.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 3.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 3.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 3.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 6.2 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.7 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 1.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.5 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 2.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 12.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 6.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 3.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 6.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 3.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 4.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.5 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 1.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 2.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 4.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 3.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 4.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |