Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
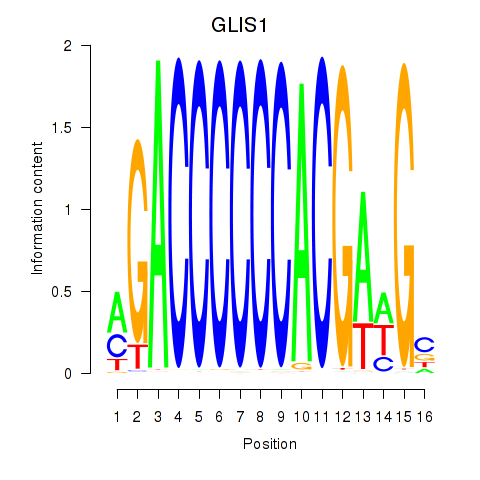
Results for GLIS1
Z-value: 0.29
Transcription factors associated with GLIS1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS1
|
ENSG00000174332.5 | GLIS family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLIS1 | hg38_v1_chr1_-_53738024_53738106 | 0.50 | 3.9e-03 | Click! |
Activity profile of GLIS1 motif
Sorted Z-values of GLIS1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_107044689 | 0.80 |
ENST00000265717.5
|
PRKAR2B
|
protein kinase cAMP-dependent type II regulatory subunit beta |
| chr18_+_11751494 | 0.57 |
ENST00000269162.9
|
GNAL
|
G protein subunit alpha L |
| chr18_+_11751467 | 0.54 |
ENST00000535121.5
|
GNAL
|
G protein subunit alpha L |
| chr11_+_68903849 | 0.51 |
ENST00000675615.1
ENST00000255078.8 |
IGHMBP2
|
immunoglobulin mu DNA binding protein 2 |
| chr19_-_49325181 | 0.51 |
ENST00000454748.7
ENST00000335875.9 ENST00000598828.1 |
SLC6A16
|
solute carrier family 6 member 16 |
| chr18_-_63158208 | 0.49 |
ENST00000678301.1
|
BCL2
|
BCL2 apoptosis regulator |
| chr18_+_11752041 | 0.39 |
ENST00000423027.8
|
GNAL
|
G protein subunit alpha L |
| chr11_+_68684534 | 0.32 |
ENST00000265643.4
|
GAL
|
galanin and GMAP prepropeptide |
| chr19_-_37906588 | 0.25 |
ENST00000447313.7
|
WDR87
|
WD repeat domain 87 |
| chr11_-_68903796 | 0.19 |
ENST00000362034.7
|
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr19_+_40191410 | 0.17 |
ENST00000253055.8
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr16_+_58025745 | 0.16 |
ENST00000219271.4
|
MMP15
|
matrix metallopeptidase 15 |
| chr19_-_41959266 | 0.07 |
ENST00000600292.5
ENST00000601078.5 ENST00000601891.5 ENST00000222008.11 |
RABAC1
|
Rab acceptor 1 |
| chr19_+_1275508 | 0.06 |
ENST00000409293.6
|
FAM174C
|
family with sequence similarity 174 member C |
Network of associatons between targets according to the STRING database.
First level regulatory network of GLIS1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 | 0.3 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.1 | 0.8 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 1.5 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 1.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |