Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
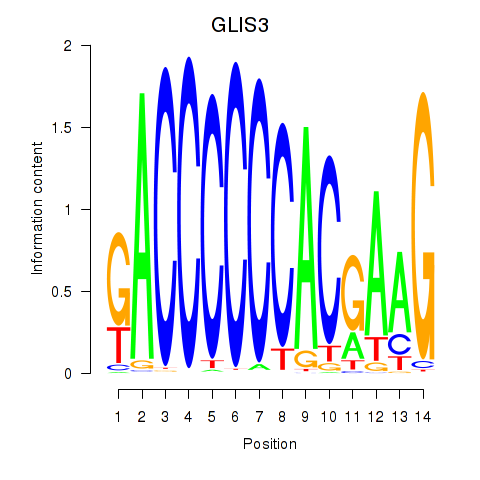
Results for GLIS3
Z-value: 0.69
Transcription factors associated with GLIS3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS3
|
ENSG00000107249.24 | GLIS family zinc finger 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLIS3 | hg38_v1_chr9_-_4299873_4299916 | -0.19 | 2.9e-01 | Click! |
Activity profile of GLIS3 motif
Sorted Z-values of GLIS3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_31413806 | 1.40 |
ENST00000536384.2
|
SERINC2
|
serine incorporator 2 |
| chr12_+_109139397 | 1.11 |
ENST00000377854.9
ENST00000377848.7 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr21_+_36699100 | 1.03 |
ENST00000290399.11
|
SIM2
|
SIM bHLH transcription factor 2 |
| chr19_-_10503186 | 0.97 |
ENST00000592055.2
ENST00000171111.10 |
KEAP1
|
kelch like ECH associated protein 1 |
| chr18_-_63158208 | 0.93 |
ENST00000678301.1
|
BCL2
|
BCL2 apoptosis regulator |
| chr12_+_54008961 | 0.90 |
ENST00000040584.6
|
HOXC8
|
homeobox C8 |
| chr11_+_68684534 | 0.88 |
ENST00000265643.4
|
GAL
|
galanin and GMAP prepropeptide |
| chr17_+_4771878 | 0.87 |
ENST00000270560.4
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr20_+_21126074 | 0.85 |
ENST00000619189.5
|
KIZ
|
kizuna centrosomal protein |
| chr20_+_21126037 | 0.83 |
ENST00000611685.4
ENST00000616848.4 |
KIZ
|
kizuna centrosomal protein |
| chr19_-_10502745 | 0.81 |
ENST00000393623.6
|
KEAP1
|
kelch like ECH associated protein 1 |
| chr13_-_20192928 | 0.79 |
ENST00000382848.5
|
GJB2
|
gap junction protein beta 2 |
| chr20_+_21125981 | 0.78 |
ENST00000619574.4
|
KIZ
|
kizuna centrosomal protein |
| chr20_+_21125999 | 0.77 |
ENST00000620891.4
|
KIZ
|
kizuna centrosomal protein |
| chr1_+_107056656 | 0.69 |
ENST00000370078.2
|
PRMT6
|
protein arginine methyltransferase 6 |
| chr3_+_127629161 | 0.68 |
ENST00000342480.7
|
PODXL2
|
podocalyxin like 2 |
| chrX_-_126552801 | 0.67 |
ENST00000371126.3
|
DCAF12L1
|
DDB1 and CUL4 associated factor 12 like 1 |
| chr16_+_89620584 | 0.66 |
ENST00000393092.7
|
DPEP1
|
dipeptidase 1 |
| chr15_-_43590155 | 0.65 |
ENST00000453080.5
ENST00000360135.8 ENST00000360301.8 ENST00000417085.2 ENST00000431962.1 ENST00000334933.8 ENST00000381879.8 ENST00000420765.5 |
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr15_-_90935130 | 0.63 |
ENST00000646620.1
|
HDDC3
|
HD domain containing 3 |
| chr14_+_104724221 | 0.59 |
ENST00000330877.7
|
ADSS1
|
adenylosuccinate synthase 1 |
| chr4_+_163343882 | 0.58 |
ENST00000338566.8
|
NPY5R
|
neuropeptide Y receptor Y5 |
| chr18_+_11752041 | 0.57 |
ENST00000423027.8
|
GNAL
|
G protein subunit alpha L |
| chr15_+_90935277 | 0.57 |
ENST00000418476.2
|
UNC45A
|
unc-45 myosin chaperone A |
| chr15_+_83447411 | 0.52 |
ENST00000324537.5
|
SH3GL3
|
SH3 domain containing GRB2 like 3, endophilin A3 |
| chr22_+_22644475 | 0.52 |
ENST00000618722.4
ENST00000652219.1 ENST00000480559.6 ENST00000448514.2 ENST00000652249.1 ENST00000651213.1 |
GGTLC2
|
gamma-glutamyltransferase light chain 2 |
| chr11_+_68903849 | 0.51 |
ENST00000675615.1
ENST00000255078.8 |
IGHMBP2
|
immunoglobulin mu DNA binding protein 2 |
| chr19_-_10502685 | 0.50 |
ENST00000591039.1
ENST00000591419.2 |
KEAP1
|
kelch like ECH associated protein 1 |
| chr17_+_47941562 | 0.49 |
ENST00000225573.5
ENST00000434554.7 ENST00000642017.2 |
PNPO
|
pyridoxamine 5'-phosphate oxidase |
| chr18_+_11751494 | 0.49 |
ENST00000269162.9
|
GNAL
|
G protein subunit alpha L |
| chr9_-_95516959 | 0.49 |
ENST00000437951.6
ENST00000430669.6 ENST00000468211.6 |
PTCH1
|
patched 1 |
| chr12_+_52949107 | 0.48 |
ENST00000388835.4
|
KRT18
|
keratin 18 |
| chr20_-_46089905 | 0.47 |
ENST00000372291.3
ENST00000290231.11 |
NCOA5
|
nuclear receptor coactivator 5 |
| chr18_+_11751467 | 0.46 |
ENST00000535121.5
|
GNAL
|
G protein subunit alpha L |
| chr5_+_172959416 | 0.45 |
ENST00000265100.6
ENST00000519239.5 |
RPL26L1
|
ribosomal protein L26 like 1 |
| chrX_+_119236245 | 0.44 |
ENST00000535419.2
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr17_+_47941506 | 0.44 |
ENST00000583599.6
|
PNPO
|
pyridoxamine 5'-phosphate oxidase |
| chr8_-_29263063 | 0.42 |
ENST00000524189.6
|
KIF13B
|
kinesin family member 13B |
| chrX_+_119236274 | 0.42 |
ENST00000217971.8
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr3_-_25783434 | 0.41 |
ENST00000396649.7
ENST00000280700.10 ENST00000428257.5 |
NGLY1
|
N-glycanase 1 |
| chr16_-_90008988 | 0.40 |
ENST00000568662.2
|
DBNDD1
|
dysbindin domain containing 1 |
| chr1_-_19799872 | 0.40 |
ENST00000294543.11
|
TMCO4
|
transmembrane and coiled-coil domains 4 |
| chr7_+_107044689 | 0.40 |
ENST00000265717.5
|
PRKAR2B
|
protein kinase cAMP-dependent type II regulatory subunit beta |
| chr11_-_68903796 | 0.39 |
ENST00000362034.7
|
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr17_+_47941694 | 0.35 |
ENST00000584061.6
|
PNPO
|
pyridoxamine 5'-phosphate oxidase |
| chr16_-_3235143 | 0.34 |
ENST00000414144.7
ENST00000431561.7 ENST00000396870.8 |
ZNF200
|
zinc finger protein 200 |
| chr17_-_35119733 | 0.34 |
ENST00000460118.6
ENST00000335858.11 |
RAD51D
|
RAD51 paralog D |
| chr6_+_33204645 | 0.33 |
ENST00000374662.4
|
HSD17B8
|
hydroxysteroid 17-beta dehydrogenase 8 |
| chr6_+_24494939 | 0.32 |
ENST00000348925.2
ENST00000357578.8 |
ALDH5A1
|
aldehyde dehydrogenase 5 family member A1 |
| chr17_+_82860354 | 0.32 |
ENST00000576996.5
|
TBCD
|
tubulin folding cofactor D |
| chr1_-_160285120 | 0.29 |
ENST00000368072.10
|
PEX19
|
peroxisomal biogenesis factor 19 |
| chr3_-_25783381 | 0.29 |
ENST00000308710.9
ENST00000676225.1 |
NGLY1
|
N-glycanase 1 |
| chr8_-_29263087 | 0.29 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr19_+_11798514 | 0.28 |
ENST00000323169.10
ENST00000450087.1 |
ZNF491
|
zinc finger protein 491 |
| chr9_-_113299196 | 0.27 |
ENST00000441031.3
|
RNF183
|
ring finger protein 183 |
| chr17_-_79952007 | 0.26 |
ENST00000574241.6
|
TBC1D16
|
TBC1 domain family member 16 |
| chr17_-_35119801 | 0.26 |
ENST00000592577.5
ENST00000590016.5 ENST00000345365.11 |
RAD51D
|
RAD51 paralog D |
| chr2_-_85602681 | 0.24 |
ENST00000334462.10
ENST00000306353.7 |
TMEM150A
|
transmembrane protein 150A |
| chr3_+_173398438 | 0.24 |
ENST00000457714.5
|
NLGN1
|
neuroligin 1 |
| chr5_-_132777371 | 0.23 |
ENST00000620483.4
|
SEPTIN8
|
septin 8 |
| chr8_+_11769639 | 0.22 |
ENST00000436750.7
|
NEIL2
|
nei like DNA glycosylase 2 |
| chr1_-_45522870 | 0.22 |
ENST00000424390.2
|
PRDX1
|
peroxiredoxin 1 |
| chr8_+_117135259 | 0.22 |
ENST00000519688.5
|
SLC30A8
|
solute carrier family 30 member 8 |
| chr2_-_85602351 | 0.21 |
ENST00000409668.1
|
TMEM150A
|
transmembrane protein 150A |
| chr17_+_50835578 | 0.21 |
ENST00000311378.5
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr1_-_53945584 | 0.20 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr2_-_240820205 | 0.20 |
ENST00000647731.1
ENST00000648680.1 ENST00000649306.1 ENST00000648129.1 ENST00000498729.9 ENST00000320389.12 ENST00000648364.1 ENST00000647885.1 ENST00000404283.9 ENST00000649096.1 |
KIF1A
|
kinesin family member 1A |
| chr19_-_37906646 | 0.20 |
ENST00000303868.5
|
WDR87
|
WD repeat domain 87 |
| chr5_-_132777344 | 0.20 |
ENST00000378706.5
|
SEPTIN8
|
septin 8 |
| chr16_+_69187125 | 0.19 |
ENST00000336278.8
|
SNTB2
|
syntrophin beta 2 |
| chr19_+_13764502 | 0.19 |
ENST00000040663.8
ENST00000319545.12 |
MRI1
|
methylthioribose-1-phosphate isomerase 1 |
| chr1_-_53945567 | 0.18 |
ENST00000371378.6
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr1_-_42335189 | 0.18 |
ENST00000361776.5
ENST00000445886.5 ENST00000361346.6 |
FOXJ3
|
forkhead box J3 |
| chr19_-_37906588 | 0.17 |
ENST00000447313.7
|
WDR87
|
WD repeat domain 87 |
| chr5_-_132777404 | 0.17 |
ENST00000296873.11
|
SEPTIN8
|
septin 8 |
| chr19_+_41354145 | 0.16 |
ENST00000604123.5
|
TMEM91
|
transmembrane protein 91 |
| chr10_+_22321056 | 0.16 |
ENST00000376663.8
|
BMI1
|
BMI1 proto-oncogene, polycomb ring finger |
| chr19_-_55580829 | 0.16 |
ENST00000592239.1
ENST00000325421.7 |
ZNF579
|
zinc finger protein 579 |
| chr19_-_14114156 | 0.16 |
ENST00000589994.6
|
PRKACA
|
protein kinase cAMP-activated catalytic subunit alpha |
| chr5_+_41925223 | 0.16 |
ENST00000296812.6
ENST00000281623.8 ENST00000509134.1 |
FBXO4
|
F-box protein 4 |
| chr9_+_129142016 | 0.14 |
ENST00000434095.2
|
PTPA
|
protein phosphatase 2 phosphatase activator |
| chr17_-_5111836 | 0.14 |
ENST00000575898.5
|
ZNF232
|
zinc finger protein 232 |
| chr8_+_117134989 | 0.13 |
ENST00000456015.7
|
SLC30A8
|
solute carrier family 30 member 8 |
| chr10_-_125161019 | 0.13 |
ENST00000411419.6
|
CTBP2
|
C-terminal binding protein 2 |
| chr19_+_1275508 | 0.13 |
ENST00000409293.6
|
FAM174C
|
family with sequence similarity 174 member C |
| chr5_-_132777215 | 0.12 |
ENST00000458488.2
|
SEPTIN8
|
septin 8 |
| chr10_-_74163 | 0.11 |
ENST00000564130.2
|
TUBB8
|
tubulin beta 8 class VIII |
| chr8_+_117135020 | 0.11 |
ENST00000518396.5
|
SLC30A8
|
solute carrier family 30 member 8 |
| chr7_+_151061217 | 0.11 |
ENST00000482950.5
ENST00000463414.5 |
SLC4A2
|
solute carrier family 4 member 2 |
| chr1_-_42335869 | 0.11 |
ENST00000372573.5
|
FOXJ3
|
forkhead box J3 |
| chr6_+_24494839 | 0.11 |
ENST00000491546.5
|
ALDH5A1
|
aldehyde dehydrogenase 5 family member A1 |
| chr16_-_1095647 | 0.10 |
ENST00000621771.1
|
C1QTNF8
|
C1q and TNF related 8 |
| chr8_-_73878816 | 0.10 |
ENST00000602593.6
ENST00000651945.1 ENST00000419880.7 ENST00000517608.5 ENST00000650817.1 |
UBE2W
|
ubiquitin conjugating enzyme E2 W |
| chr17_-_81959769 | 0.09 |
ENST00000477214.5
|
NOTUM
|
notum, palmitoleoyl-protein carboxylesterase |
| chr12_-_47758828 | 0.09 |
ENST00000389212.7
ENST00000449771.7 |
RAPGEF3
|
Rap guanine nucleotide exchange factor 3 |
| chr9_-_136764515 | 0.09 |
ENST00000316144.6
|
LCN15
|
lipocalin 15 |
| chr8_+_11769696 | 0.09 |
ENST00000455213.6
ENST00000403422.7 ENST00000528323.5 ENST00000284503.7 |
NEIL2
|
nei like DNA glycosylase 2 |
| chr17_+_47941721 | 0.08 |
ENST00000641511.1
|
PNPO
|
pyridoxamine 5'-phosphate oxidase |
| chr17_-_3916455 | 0.08 |
ENST00000225538.4
|
P2RX1
|
purinergic receptor P2X 1 |
| chr18_+_3451647 | 0.08 |
ENST00000345133.9
ENST00000330513.10 ENST00000549546.5 |
TGIF1
|
TGFB induced factor homeobox 1 |
| chr17_+_50834581 | 0.07 |
ENST00000426127.1
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr5_-_132777866 | 0.06 |
ENST00000448933.5
|
SEPTIN8
|
septin 8 |
| chr11_+_818906 | 0.06 |
ENST00000336615.9
|
PNPLA2
|
patatin like phospholipase domain containing 2 |
| chr10_-_125161056 | 0.06 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr1_-_45521854 | 0.04 |
ENST00000372079.1
ENST00000319248.13 |
PRDX1
|
peroxiredoxin 1 |
| chr15_-_43589942 | 0.04 |
ENST00000429176.5
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr7_+_151061499 | 0.04 |
ENST00000488420.1
|
SLC4A2
|
solute carrier family 4 member 2 |
| chr9_+_129141637 | 0.02 |
ENST00000419582.5
|
PTPA
|
protein phosphatase 2 phosphatase activator |
| chr9_+_129141954 | 0.02 |
ENST00000432651.5
ENST00000435132.5 |
PTPA
|
protein phosphatase 2 phosphatase activator |
| chr6_+_44223770 | 0.01 |
ENST00000652453.1
ENST00000393841.6 ENST00000371724.6 ENST00000642777.1 ENST00000645692.1 |
SLC29A1
|
solute carrier family 29 member 1 (Augustine blood group) |
| chr9_-_136304084 | 0.00 |
ENST00000638797.2
ENST00000624277.3 |
CCDC187
|
coiled-coil domain containing 187 |
| chr18_+_3451585 | 0.00 |
ENST00000551541.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GLIS3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.2 | 0.7 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.2 | 1.4 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) |
| 0.2 | 0.7 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.2 | 0.9 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.2 | 0.9 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.2 | 0.5 | GO:0043000 | Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.2 | 1.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 2.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.8 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.1 | 0.6 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.1 | 0.5 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.6 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.4 | GO:0006540 | acetate metabolic process(GO:0006083) glutamate decarboxylation to succinate(GO:0006540) |
| 0.1 | 0.2 | GO:0048789 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.1 | 0.7 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 1.5 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.5 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.5 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.3 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.2 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.0 | 0.7 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.1 | GO:0002442 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.3 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.8 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.5 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.9 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 2.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0004733 | pyridoxamine-phosphate oxidase activity(GO:0004733) |
| 0.2 | 0.7 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.2 | 0.6 | GO:0008893 | guanosine-3',5'-bis(diphosphate) 3'-diphosphatase activity(GO:0008893) diphosphoric monoester hydrolase activity(GO:0016794) |
| 0.2 | 0.7 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.2 | 1.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.6 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.1 | 0.9 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.3 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 1.4 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.7 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.7 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.5 | GO:0005119 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 1.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |