Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
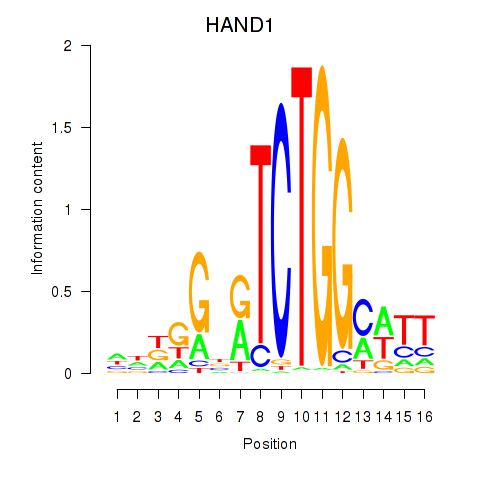
Results for HAND1
Z-value: 1.03
Transcription factors associated with HAND1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HAND1
|
ENSG00000113196.3 | heart and neural crest derivatives expressed 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HAND1 | hg38_v1_chr5_-_154478218_154478234 | -0.18 | 3.3e-01 | Click! |
Activity profile of HAND1 motif
Sorted Z-values of HAND1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_52803807 | 4.02 |
ENST00000334575.6
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr14_-_64942720 | 3.37 |
ENST00000557049.1
ENST00000389614.6 |
GPX2
|
glutathione peroxidase 2 |
| chr6_+_133889105 | 3.13 |
ENST00000367882.5
|
TCF21
|
transcription factor 21 |
| chr14_-_64942783 | 2.76 |
ENST00000612794.1
|
GPX2
|
glutathione peroxidase 2 |
| chr2_-_27495185 | 2.75 |
ENST00000264703.4
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr16_-_31135699 | 2.66 |
ENST00000317508.11
ENST00000568261.5 ENST00000567797.1 |
PRSS8
|
serine protease 8 |
| chr17_-_66220630 | 2.66 |
ENST00000585162.1
|
APOH
|
apolipoprotein H |
| chr12_+_40692413 | 2.38 |
ENST00000551295.7
ENST00000547702.5 ENST00000551424.5 |
CNTN1
|
contactin 1 |
| chr17_+_47831608 | 2.38 |
ENST00000269025.9
|
LRRC46
|
leucine rich repeat containing 46 |
| chr2_+_209579598 | 2.17 |
ENST00000445941.5
ENST00000673860.1 |
MAP2
|
microtubule associated protein 2 |
| chr2_+_209579399 | 2.14 |
ENST00000360351.8
|
MAP2
|
microtubule associated protein 2 |
| chr2_+_227813834 | 2.13 |
ENST00000358813.5
ENST00000409189.7 |
CCL20
|
C-C motif chemokine ligand 20 |
| chr7_-_81770039 | 1.96 |
ENST00000222390.11
ENST00000453411.6 ENST00000457544.7 ENST00000444829.7 |
HGF
|
hepatocyte growth factor |
| chr11_-_5243644 | 1.91 |
ENST00000643122.1
|
HBD
|
hemoglobin subunit delta |
| chr20_+_59721210 | 1.91 |
ENST00000395636.6
ENST00000361300.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr17_+_39628496 | 1.90 |
ENST00000394265.5
ENST00000394267.2 |
PPP1R1B
|
protein phosphatase 1 regulatory inhibitor subunit 1B |
| chr19_+_35868585 | 1.90 |
ENST00000652533.1
|
APLP1
|
amyloid beta precursor like protein 1 |
| chr19_+_35868518 | 1.88 |
ENST00000221891.9
|
APLP1
|
amyloid beta precursor like protein 1 |
| chr7_-_117323041 | 1.66 |
ENST00000491214.1
ENST00000265441.8 |
WNT2
|
Wnt family member 2 |
| chr11_-_63563370 | 1.56 |
ENST00000255695.2
|
PLAAT2
|
phospholipase A and acyltransferase 2 |
| chr2_+_209579429 | 1.51 |
ENST00000361559.8
|
MAP2
|
microtubule associated protein 2 |
| chr1_+_202122910 | 1.50 |
ENST00000682545.1
ENST00000367282.6 ENST00000682887.1 |
GPR37L1
|
G protein-coupled receptor 37 like 1 |
| chr20_+_46029165 | 1.40 |
ENST00000616201.4
ENST00000616202.4 ENST00000616933.4 ENST00000626937.2 |
SLC12A5
|
solute carrier family 12 member 5 |
| chr7_-_81770122 | 1.39 |
ENST00000423064.7
|
HGF
|
hepatocyte growth factor |
| chr2_+_209579315 | 1.35 |
ENST00000392193.5
|
MAP2
|
microtubule associated protein 2 |
| chr16_-_31135640 | 1.33 |
ENST00000567531.5
|
PRSS8
|
serine protease 8 |
| chr7_+_73827737 | 1.29 |
ENST00000435050.1
|
CLDN4
|
claudin 4 |
| chr19_-_10102659 | 1.27 |
ENST00000592641.5
ENST00000253109.5 |
ANGPTL6
|
angiopoietin like 6 |
| chr9_-_97938157 | 1.26 |
ENST00000616898.2
|
HEMGN
|
hemogen |
| chr11_-_62709633 | 1.23 |
ENST00000532818.5
|
BSCL2
|
BSCL2 lipid droplet biogenesis associated, seipin |
| chr6_-_132798587 | 1.23 |
ENST00000275227.9
|
SLC18B1
|
solute carrier family 18 member B1 |
| chr1_+_27392612 | 1.22 |
ENST00000374024.4
|
GPR3
|
G protein-coupled receptor 3 |
| chr1_+_65309517 | 1.22 |
ENST00000371069.5
|
DNAJC6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr7_-_81770013 | 1.20 |
ENST00000465234.2
|
HGF
|
hepatocyte growth factor |
| chr1_+_207089233 | 1.20 |
ENST00000243611.9
ENST00000367076.7 |
C4BPB
|
complement component 4 binding protein beta |
| chr15_+_74217189 | 1.15 |
ENST00000635913.1
|
CCDC33
|
coiled-coil domain containing 33 |
| chr1_+_202122881 | 1.13 |
ENST00000683302.1
ENST00000683557.1 |
GPR37L1
|
G protein-coupled receptor 37 like 1 |
| chr22_+_24603147 | 1.13 |
ENST00000412658.5
ENST00000445029.5 ENST00000400382.6 ENST00000419133.5 ENST00000438643.6 ENST00000452551.5 ENST00000412898.5 ENST00000400380.5 ENST00000455483.5 ENST00000430289.5 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr8_+_30744150 | 1.12 |
ENST00000265616.10
ENST00000341403.9 ENST00000615729.4 |
UBXN8
|
UBX domain protein 8 |
| chr11_-_62709841 | 1.10 |
ENST00000464544.1
ENST00000530009.1 |
BSCL2
|
BSCL2 lipid droplet biogenesis associated, seipin |
| chr1_+_207089195 | 1.08 |
ENST00000452902.6
|
C4BPB
|
complement component 4 binding protein beta |
| chr3_+_112333147 | 1.05 |
ENST00000473539.5
ENST00000315711.12 ENST00000383681.7 |
CD200
|
CD200 molecule |
| chr1_+_207088825 | 1.04 |
ENST00000367078.8
|
C4BPB
|
complement component 4 binding protein beta |
| chrX_+_71144818 | 1.04 |
ENST00000536169.5
ENST00000358741.4 ENST00000395855.6 ENST00000374051.7 |
NLGN3
|
neuroligin 3 |
| chr1_+_207089283 | 1.03 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein beta |
| chr19_+_4304588 | 0.95 |
ENST00000221856.11
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr4_+_41612892 | 0.93 |
ENST00000509454.5
ENST00000396595.7 ENST00000381753.8 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr7_-_81769971 | 0.89 |
ENST00000354224.10
ENST00000643024.1 |
HGF
|
hepatocyte growth factor |
| chr6_+_43132472 | 0.84 |
ENST00000489707.5
|
PTK7
|
protein tyrosine kinase 7 (inactive) |
| chr11_+_101914997 | 0.83 |
ENST00000263468.13
|
CEP126
|
centrosomal protein 126 |
| chr2_-_196926670 | 0.83 |
ENST00000354764.9
|
PGAP1
|
post-GPI attachment to proteins inositol deacylase 1 |
| chr10_-_71719772 | 0.81 |
ENST00000441508.4
|
C10orf105
|
chromosome 10 open reading frame 105 |
| chr15_-_76012390 | 0.80 |
ENST00000394907.8
|
NRG4
|
neuregulin 4 |
| chr7_-_20217342 | 0.80 |
ENST00000400331.10
ENST00000332878.8 |
MACC1
|
MET transcriptional regulator MACC1 |
| chr10_+_125973373 | 0.80 |
ENST00000417114.5
ENST00000445510.5 ENST00000368691.5 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr16_-_28597042 | 0.77 |
ENST00000533150.5
ENST00000335715.9 |
SULT1A2
|
sulfotransferase family 1A member 2 |
| chr3_+_112332500 | 0.77 |
ENST00000606471.5
|
CD200
|
CD200 molecule |
| chr18_+_58045642 | 0.76 |
ENST00000676223.1
ENST00000675147.1 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr1_-_101996919 | 0.75 |
ENST00000370103.9
|
OLFM3
|
olfactomedin 3 |
| chr7_-_151057848 | 0.75 |
ENST00000297518.4
|
CDK5
|
cyclin dependent kinase 5 |
| chr7_-_81770428 | 0.75 |
ENST00000421558.1
|
HGF
|
hepatocyte growth factor |
| chr16_-_28610032 | 0.74 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chr3_+_130581741 | 0.74 |
ENST00000511332.1
|
COL6A6
|
collagen type VI alpha 6 chain |
| chr12_-_62260123 | 0.70 |
ENST00000552075.5
|
TAFA2
|
TAFA chemokine like family member 2 |
| chr12_-_25648544 | 0.70 |
ENST00000540106.5
ENST00000445693.5 ENST00000545543.1 |
LMNTD1
|
lamin tail domain containing 1 |
| chr16_-_28609992 | 0.69 |
ENST00000314752.11
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chr2_-_158380960 | 0.68 |
ENST00000409187.5
|
CCDC148
|
coiled-coil domain containing 148 |
| chr6_+_123803853 | 0.68 |
ENST00000368417.6
|
NKAIN2
|
sodium/potassium transporting ATPase interacting 2 |
| chr12_-_52814106 | 0.67 |
ENST00000551956.2
|
KRT4
|
keratin 4 |
| chr18_+_58045683 | 0.67 |
ENST00000592846.5
ENST00000675801.1 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chrX_+_150363306 | 0.66 |
ENST00000370401.7
ENST00000432680.7 |
MAMLD1
|
mastermind like domain containing 1 |
| chr3_+_46877705 | 0.66 |
ENST00000449590.6
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr20_-_1184981 | 0.66 |
ENST00000429036.2
|
TMEM74B
|
transmembrane protein 74B |
| chr7_-_151057880 | 0.65 |
ENST00000485972.6
|
CDK5
|
cyclin dependent kinase 5 |
| chr16_-_28609976 | 0.64 |
ENST00000566189.5
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chr7_-_81770362 | 0.64 |
ENST00000412881.5
|
HGF
|
hepatocyte growth factor |
| chr3_-_9769910 | 0.62 |
ENST00000256460.8
|
CAMK1
|
calcium/calmodulin dependent protein kinase I |
| chr6_-_39314412 | 0.59 |
ENST00000373231.9
|
KCNK17
|
potassium two pore domain channel subfamily K member 17 |
| chr6_+_11093753 | 0.59 |
ENST00000416247.4
|
SMIM13
|
small integral membrane protein 13 |
| chr17_-_74623730 | 0.59 |
ENST00000392619.2
|
CD300E
|
CD300e molecule |
| chr19_+_38304105 | 0.57 |
ENST00000588605.5
ENST00000301246.10 |
C19orf33
|
chromosome 19 open reading frame 33 |
| chr5_+_36608146 | 0.56 |
ENST00000381918.4
ENST00000513646.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr10_-_102451536 | 0.56 |
ENST00000625129.1
|
C10orf95
|
chromosome 10 open reading frame 95 |
| chr4_-_40629842 | 0.55 |
ENST00000295971.12
|
RBM47
|
RNA binding motif protein 47 |
| chr7_+_99408609 | 0.55 |
ENST00000403633.6
|
BUD31
|
BUD31 homolog |
| chr10_+_5048748 | 0.54 |
ENST00000602997.5
ENST00000439082.7 |
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr7_-_142885737 | 0.53 |
ENST00000359396.9
ENST00000436401.1 |
TRPV6
|
transient receptor potential cation channel subfamily V member 6 |
| chrX_+_150363258 | 0.52 |
ENST00000683696.1
|
MAMLD1
|
mastermind like domain containing 1 |
| chr2_-_42361149 | 0.49 |
ENST00000468711.5
ENST00000463055.1 |
COX7A2L
|
cytochrome c oxidase subunit 7A2 like |
| chr2_-_196926717 | 0.49 |
ENST00000409475.5
ENST00000374738.3 |
PGAP1
|
post-GPI attachment to proteins inositol deacylase 1 |
| chr18_-_46756791 | 0.49 |
ENST00000538168.5
ENST00000536490.1 |
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr4_+_25160631 | 0.45 |
ENST00000510415.1
ENST00000507794.2 ENST00000512921.4 |
SEPSECS-AS1
PI4K2B
|
SEPSECS antisense RNA 1 (head to head) phosphatidylinositol 4-kinase type 2 beta |
| chr4_-_25160375 | 0.45 |
ENST00000382103.7
|
SEPSECS
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr1_+_174877430 | 0.45 |
ENST00000392064.6
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr17_+_38428456 | 0.44 |
ENST00000622683.5
ENST00000620417.4 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr4_+_41612702 | 0.42 |
ENST00000509277.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr7_+_111091006 | 0.42 |
ENST00000451085.5
ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr16_-_30031259 | 0.42 |
ENST00000380495.8
|
TLCD3B
|
TLC domain containing 3B |
| chr7_+_111091119 | 0.42 |
ENST00000308478.10
|
LRRN3
|
leucine rich repeat neuronal 3 |
| chr3_+_147393718 | 0.42 |
ENST00000488404.5
|
ZIC1
|
Zic family member 1 |
| chr20_+_46029009 | 0.42 |
ENST00000608944.5
|
SLC12A5
|
solute carrier family 12 member 5 |
| chr1_-_167914089 | 0.42 |
ENST00000476818.2
ENST00000367848.1 ENST00000367851.9 ENST00000545172.5 |
ADCY10
|
adenylate cyclase 10 |
| chr10_+_97584314 | 0.41 |
ENST00000370647.8
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr6_-_39314445 | 0.40 |
ENST00000453413.2
|
KCNK17
|
potassium two pore domain channel subfamily K member 17 |
| chr4_-_40630588 | 0.38 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr17_+_45844875 | 0.36 |
ENST00000329196.7
|
SPPL2C
|
signal peptide peptidase like 2C |
| chr19_+_507487 | 0.36 |
ENST00000359315.6
|
TPGS1
|
tubulin polyglutamylase complex subunit 1 |
| chr6_+_123804141 | 0.36 |
ENST00000368416.5
|
NKAIN2
|
sodium/potassium transporting ATPase interacting 2 |
| chr1_-_53940100 | 0.36 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr14_+_73616844 | 0.35 |
ENST00000381139.1
|
ACOT6
|
acyl-CoA thioesterase 6 |
| chr5_+_144205250 | 0.33 |
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16 |
| chr14_-_74426102 | 0.33 |
ENST00000554953.1
ENST00000331628.8 |
SYNDIG1L
|
synapse differentiation inducing 1 like |
| chr10_+_132036652 | 0.32 |
ENST00000657785.1
|
JAKMIP3
|
Janus kinase and microtubule interacting protein 3 |
| chr2_+_201132769 | 0.32 |
ENST00000494258.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr16_-_65072207 | 0.30 |
ENST00000562882.5
ENST00000567934.5 |
CDH11
|
cadherin 11 |
| chr7_+_112450451 | 0.29 |
ENST00000429071.5
ENST00000403825.8 ENST00000675268.1 |
IFRD1
ENSG00000288634.1
|
interferon related developmental regulator 1 novel protein |
| chrX_-_34657274 | 0.29 |
ENST00000275954.4
|
TMEM47
|
transmembrane protein 47 |
| chr12_-_56360084 | 0.29 |
ENST00000314128.9
ENST00000557235.5 ENST00000651915.1 |
STAT2
|
signal transducer and activator of transcription 2 |
| chr4_-_103019634 | 0.28 |
ENST00000510559.1
ENST00000296422.12 ENST00000394789.7 |
SLC9B1
|
solute carrier family 9 member B1 |
| chr6_-_36986122 | 0.28 |
ENST00000460219.2
ENST00000373627.10 ENST00000373616.9 |
MTCH1
|
mitochondrial carrier 1 |
| chr4_-_25160546 | 0.27 |
ENST00000681948.1
|
SEPSECS
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr18_-_46757012 | 0.27 |
ENST00000315087.12
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr9_-_120793377 | 0.27 |
ENST00000684001.1
ENST00000684405.1 ENST00000608872.6 |
FBXW2
|
F-box and WD repeat domain containing 2 |
| chr7_+_76510516 | 0.27 |
ENST00000257632.9
|
UPK3B
|
uroplakin 3B |
| chr12_+_25958891 | 0.26 |
ENST00000381352.7
ENST00000535907.5 ENST00000405154.6 ENST00000615708.4 |
RASSF8
|
Ras association domain family member 8 |
| chr16_+_90022600 | 0.24 |
ENST00000620723.4
ENST00000268699.9 |
GAS8
|
growth arrest specific 8 |
| chr17_+_7933080 | 0.23 |
ENST00000565740.5
|
CNTROB
|
centrobin, centriole duplication and spindle assembly protein |
| chrX_-_134796256 | 0.22 |
ENST00000486347.5
|
PABIR2
|
PABIR family member 2 |
| chr19_-_12834548 | 0.21 |
ENST00000590404.6
ENST00000592204.5 ENST00000674343.2 |
RTBDN
|
retbindin |
| chr14_+_104138578 | 0.20 |
ENST00000423312.7
|
KIF26A
|
kinesin family member 26A |
| chr22_+_24607658 | 0.20 |
ENST00000451366.5
ENST00000428855.5 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr19_+_44662278 | 0.20 |
ENST00000622237.4
ENST00000618372.4 ENST00000619799.4 ENST00000592789.5 ENST00000591979.1 |
CEACAM19
|
CEA cell adhesion molecule 19 |
| chr14_-_93207007 | 0.20 |
ENST00000556566.1
ENST00000306954.5 |
GON7
|
GON7 subunit of KEOPS complex |
| chr10_+_97584347 | 0.20 |
ENST00000370649.3
ENST00000370646.9 |
ENSG00000249967.1
HOGA1
|
novel protein 4-hydroxy-2-oxoglutarate aldolase 1 |
| chr9_-_120793343 | 0.19 |
ENST00000684047.1
|
FBXW2
|
F-box and WD repeat domain containing 2 |
| chr5_+_14441043 | 0.19 |
ENST00000639876.2
|
TRIO
|
trio Rho guanine nucleotide exchange factor |
| chr7_+_97005538 | 0.18 |
ENST00000518156.3
|
DLX6
|
distal-less homeobox 6 |
| chr7_+_141790217 | 0.18 |
ENST00000247883.5
|
TAS2R5
|
taste 2 receptor member 5 |
| chr19_-_40425982 | 0.17 |
ENST00000357949.5
|
SERTAD1
|
SERTA domain containing 1 |
| chr2_-_42361198 | 0.16 |
ENST00000234301.3
|
COX7A2L
|
cytochrome c oxidase subunit 7A2 like |
| chrX_-_68433301 | 0.16 |
ENST00000680804.1
ENST00000680612.1 |
OPHN1
|
oligophrenin 1 |
| chr1_-_155934398 | 0.15 |
ENST00000368320.7
ENST00000368321.8 |
KHDC4
|
KH domain containing 4, pre-mRNA splicing factor |
| chr3_+_124094663 | 0.14 |
ENST00000460856.5
ENST00000240874.7 |
KALRN
|
kalirin RhoGEF kinase |
| chr6_+_6588708 | 0.13 |
ENST00000230568.5
|
LY86
|
lymphocyte antigen 86 |
| chr22_-_39532722 | 0.13 |
ENST00000334678.8
|
RPS19BP1
|
ribosomal protein S19 binding protein 1 |
| chr6_+_101181254 | 0.11 |
ENST00000682090.1
ENST00000421544.6 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr10_-_5003850 | 0.11 |
ENST00000421196.7
ENST00000455190.2 ENST00000380753.8 |
AKR1C2
|
aldo-keto reductase family 1 member C2 |
| chr4_+_41613476 | 0.11 |
ENST00000508466.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr12_-_53335737 | 0.11 |
ENST00000303846.3
|
SP7
|
Sp7 transcription factor |
| chr1_+_43979179 | 0.10 |
ENST00000434555.7
ENST00000372324.6 ENST00000481924.2 |
B4GALT2
|
beta-1,4-galactosyltransferase 2 |
| chr14_-_68793055 | 0.10 |
ENST00000439696.3
|
ZFP36L1
|
ZFP36 ring finger protein like 1 |
| chr4_+_75556048 | 0.10 |
ENST00000616557.1
ENST00000435974.2 ENST00000311623.9 |
ODAPH
|
odontogenesis associated phosphoprotein |
| chr10_+_132036336 | 0.09 |
ENST00000668452.1
|
JAKMIP3
|
Janus kinase and microtubule interacting protein 3 |
| chr11_-_26722051 | 0.08 |
ENST00000396005.8
|
SLC5A12
|
solute carrier family 5 member 12 |
| chr10_+_101354028 | 0.08 |
ENST00000393441.8
|
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr21_+_33230073 | 0.08 |
ENST00000342101.7
ENST00000413881.5 ENST00000443073.5 |
IFNAR2
|
interferon alpha and beta receptor subunit 2 |
| chr3_+_15601736 | 0.07 |
ENST00000303498.10
ENST00000417015.3 ENST00000672065.1 ENST00000672112.1 ENST00000643237.3 ENST00000672141.1 ENST00000646371.1 ENST00000672427.1 ENST00000672336.1 ENST00000673467.1 ENST00000672760.1 ENST00000437172.6 |
BTD
|
biotinidase |
| chr21_+_33229883 | 0.06 |
ENST00000382264.7
ENST00000342136.9 ENST00000404220.7 |
IFNAR2
|
interferon alpha and beta receptor subunit 2 |
| chr6_-_87095059 | 0.06 |
ENST00000369582.6
ENST00000610310.3 ENST00000630630.2 ENST00000627148.3 ENST00000625577.1 |
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr15_-_78811415 | 0.06 |
ENST00000388820.5
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif 7 |
| chr19_+_18382163 | 0.05 |
ENST00000595973.2
|
GDF15
|
growth differentiation factor 15 |
| chr11_+_111540273 | 0.05 |
ENST00000533999.5
|
LAYN
|
layilin |
| chr20_+_31368594 | 0.05 |
ENST00000253381.3
|
DEFB118
|
defensin beta 118 |
| chr19_-_12835400 | 0.05 |
ENST00000591512.2
ENST00000587549.1 ENST00000322912.9 |
RTBDN
|
retbindin |
| chr19_+_55488404 | 0.04 |
ENST00000594321.5
ENST00000389623.11 |
SSC5D
|
scavenger receptor cysteine rich family member with 5 domains |
| chr4_-_40630739 | 0.03 |
ENST00000511902.5
ENST00000505220.1 |
RBM47
|
RNA binding motif protein 47 |
| chr3_+_124094696 | 0.03 |
ENST00000360013.7
ENST00000684186.1 ENST00000684276.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr9_+_70043840 | 0.03 |
ENST00000377182.5
|
MAMDC2
|
MAM domain containing 2 |
| chr12_+_120694167 | 0.02 |
ENST00000535656.1
|
MLEC
|
malectin |
| chr6_-_117602447 | 0.02 |
ENST00000368498.7
ENST00000052569.10 ENST00000535237.2 |
GOPC
|
golgi associated PDZ and coiled-coil motif containing |
| chr10_+_80537902 | 0.02 |
ENST00000339284.6
ENST00000646907.2 |
SH2D4B
|
SH2 domain containing 4B |
| chr2_+_230996115 | 0.01 |
ENST00000424440.5
ENST00000452881.5 ENST00000433428.6 ENST00000455816.1 ENST00000440792.5 ENST00000423134.1 |
SPATA3
|
spermatogenesis associated 3 |
| chr2_-_207167220 | 0.01 |
ENST00000421199.5
ENST00000457962.5 |
KLF7
|
Kruppel like factor 7 |
| chr1_-_155934370 | 0.00 |
ENST00000368319.3
|
KHDC4
|
KH domain containing 4, pre-mRNA splicing factor |
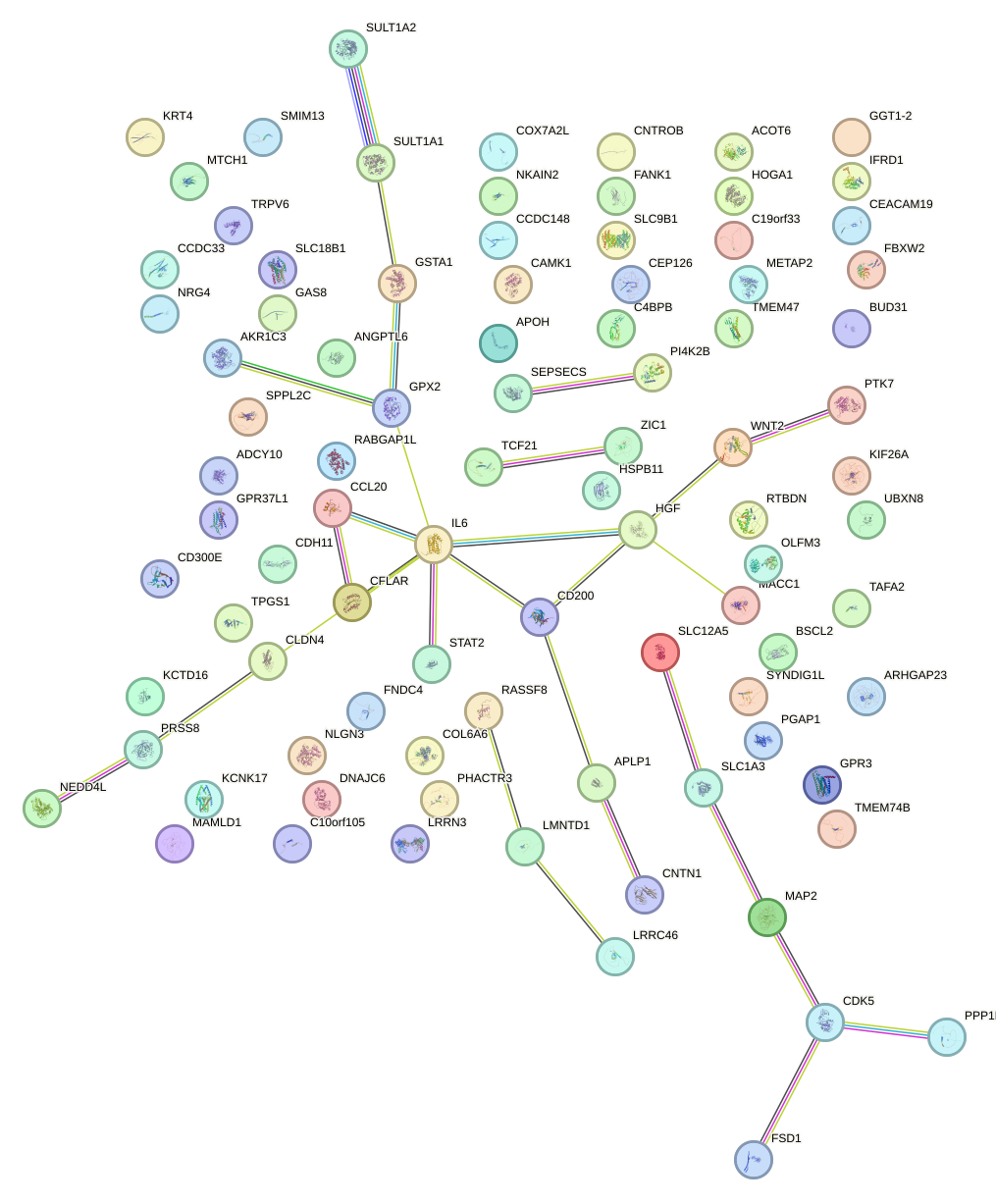
Network of associatons between targets according to the STRING database.
First level regulatory network of HAND1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.6 | 3.8 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.6 | 6.8 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.5 | 2.1 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.5 | 4.3 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.5 | 1.8 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.3 | 1.0 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.2 | 2.7 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.2 | 2.6 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.2 | 1.3 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.2 | 1.7 | GO:0060424 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.2 | 0.5 | GO:0016487 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.2 | 0.7 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.2 | 1.4 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 0.6 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 1.9 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 4.0 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 1.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 2.8 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.3 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 1.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 1.8 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 1.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 1.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 6.0 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 2.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.5 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.6 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 0.8 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 7.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.6 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 1.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 6.1 | GO:1990748 | cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 1.3 | GO:0061436 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) establishment of skin barrier(GO:0061436) |
| 0.0 | 0.1 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.2 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.5 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.7 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.2 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.4 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.1 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.8 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 2.4 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.2 | GO:0046959 | habituation(GO:0046959) negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.3 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.8 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.0 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 1.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 2.7 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 1.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 1.7 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 4.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.4 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 6.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 4.3 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 1.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 3.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 3.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0031696 | alpha-2C adrenergic receptor binding(GO:0031696) |
| 0.9 | 2.6 | GO:0036505 | prosaposin receptor activity(GO:0036505) |
| 0.5 | 2.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.5 | 1.9 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.5 | 2.8 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.5 | 1.4 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.4 | 2.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 10.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 6.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 0.5 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.1 | 0.7 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 6.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.8 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.6 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.8 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 1.7 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 1.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 5.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 3.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 1.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.0 | 0.4 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 1.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.3 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 6.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 7.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 3.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 4.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 5.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 3.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 2.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 2.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 2.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |