Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
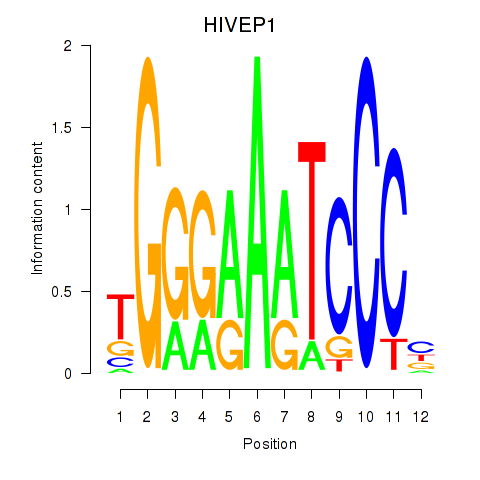
Results for HIVEP1
Z-value: 1.94
Transcription factors associated with HIVEP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIVEP1
|
ENSG00000095951.17 | HIVEP zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HIVEP1 | hg38_v1_chr6_+_12012304_12012338 | 0.42 | 1.6e-02 | Click! |
Activity profile of HIVEP1 motif
Sorted Z-values of HIVEP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_106335613 | 10.07 |
ENST00000603660.1
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr14_-_106235582 | 9.12 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr12_-_9760893 | 8.30 |
ENST00000228434.7
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr14_-_106593319 | 7.91 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr14_-_106062670 | 7.88 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr14_-_106360320 | 7.84 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
| chr14_-_105856183 | 7.84 |
ENST00000637539.2
ENST00000390559.6 |
IGHM
|
immunoglobulin heavy constant mu |
| chr14_-_106675544 | 7.06 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr14_-_106538331 | 7.01 |
ENST00000390624.3
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr14_-_106130061 | 5.53 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr16_+_32066065 | 5.21 |
ENST00000354689.6
|
IGHV3OR16-9
|
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr14_-_106154113 | 4.93 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr14_-_106108453 | 4.79 |
ENST00000632950.2
|
IGHV5-10-1
|
immunoglobulin heavy variable 5-10-1 |
| chr7_-_150800320 | 4.74 |
ENST00000492607.5
ENST00000326442.10 ENST00000450753.2 |
TMEM176B
|
transmembrane protein 176B |
| chr14_-_106088573 | 4.71 |
ENST00000632099.1
|
IGHV3-64D
|
immunoglobulin heavy variable 3-64D |
| chr14_-_106411021 | 4.66 |
ENST00000390618.2
|
IGHV3-38
|
immunoglobulin heavy variable 3-38 (non-functional) |
| chr14_-_106579223 | 4.55 |
ENST00000390626.2
|
IGHV5-51
|
immunoglobulin heavy variable 5-51 |
| chr14_-_106791226 | 4.32 |
ENST00000433072.2
|
IGHV3-72
|
immunoglobulin heavy variable 3-72 |
| chr6_+_32439866 | 3.96 |
ENST00000374982.5
ENST00000395388.7 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr6_+_32741382 | 3.92 |
ENST00000374940.4
|
HLA-DQA2
|
major histocompatibility complex, class II, DQ alpha 2 |
| chr22_-_37244237 | 3.85 |
ENST00000401529.3
ENST00000249071.11 |
RAC2
|
Rac family small GTPase 2 |
| chr14_-_106211453 | 3.83 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr22_+_36860973 | 3.69 |
ENST00000447071.5
ENST00000397147.7 ENST00000248899.11 |
NCF4
|
neutrophil cytosolic factor 4 |
| chr4_-_108166750 | 3.51 |
ENST00000515500.5
|
LEF1
|
lymphoid enhancer binding factor 1 |
| chr6_+_31587268 | 3.48 |
ENST00000396101.7
ENST00000490742.5 |
LST1
|
leukocyte specific transcript 1 |
| chr6_+_29723421 | 3.47 |
ENST00000259951.12
ENST00000434407.6 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr3_-_112133218 | 3.46 |
ENST00000488580.5
ENST00000308910.9 ENST00000460387.6 ENST00000484193.5 ENST00000487901.1 |
GCSAM
|
germinal center associated signaling and motility |
| chr6_+_31587185 | 3.46 |
ENST00000376092.7
ENST00000376086.7 ENST00000303757.12 ENST00000376093.6 |
LST1
|
leukocyte specific transcript 1 |
| chr14_-_106811131 | 3.33 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr17_+_42288464 | 3.30 |
ENST00000590726.7
ENST00000678903.1 ENST00000590949.6 ENST00000676585.1 ENST00000588868.5 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr16_+_33827140 | 3.27 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr6_-_31357171 | 3.26 |
ENST00000412585.7
ENST00000434333.1 |
HLA-B
|
major histocompatibility complex, class I, B |
| chr4_-_108166715 | 3.22 |
ENST00000510624.5
|
LEF1
|
lymphoid enhancer binding factor 1 |
| chr14_-_106803221 | 3.19 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chrX_+_47585212 | 3.18 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr7_-_150800533 | 3.10 |
ENST00000434545.5
|
TMEM176B
|
transmembrane protein 176B |
| chr4_-_74099187 | 3.08 |
ENST00000508487.3
|
CXCL2
|
C-X-C motif chemokine ligand 2 |
| chr6_+_29723340 | 3.06 |
ENST00000334668.8
|
HLA-F
|
major histocompatibility complex, class I, F |
| chr1_+_37474572 | 3.05 |
ENST00000373087.7
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr19_-_10334723 | 3.01 |
ENST00000592945.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr6_+_14117764 | 2.99 |
ENST00000379153.4
|
CD83
|
CD83 molecule |
| chr6_-_31272069 | 2.99 |
ENST00000415537.1
ENST00000376228.9 ENST00000383329.7 |
HLA-C
|
major histocompatibility complex, class I, C |
| chr5_-_39219555 | 2.98 |
ENST00000512982.4
ENST00000351578.12 ENST00000509072.5 ENST00000504542.1 ENST00000506557.5 |
FYB1
|
FYN binding protein 1 |
| chr4_-_73998669 | 2.97 |
ENST00000296027.5
|
CXCL5
|
C-X-C motif chemokine ligand 5 |
| chr14_+_75522427 | 2.95 |
ENST00000286639.8
|
BATF
|
basic leucine zipper ATF-like transcription factor |
| chr17_+_42288429 | 2.94 |
ENST00000676631.1
ENST00000677893.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr9_-_133479075 | 2.92 |
ENST00000414172.1
ENST00000371897.8 ENST00000371899.9 |
SLC2A6
|
solute carrier family 2 member 6 |
| chr22_-_37244417 | 2.88 |
ENST00000405484.5
ENST00000441619.5 ENST00000406508.5 |
RAC2
|
Rac family small GTPase 2 |
| chr19_-_46625037 | 2.87 |
ENST00000596260.1
ENST00000594275.1 ENST00000597185.1 ENST00000598865.5 ENST00000291294.7 |
PTGIR
|
prostaglandin I2 receptor |
| chr12_-_57478070 | 2.85 |
ENST00000549602.1
ENST00000430041.6 |
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr8_+_27311471 | 2.83 |
ENST00000397501.5
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr19_-_42132391 | 2.80 |
ENST00000528894.8
ENST00000560804.6 ENST00000560558.5 ENST00000560398.5 ENST00000526816.6 ENST00000625670.2 |
POU2F2
|
POU class 2 homeobox 2 |
| chr11_+_72814380 | 2.72 |
ENST00000534905.5
ENST00000321297.10 ENST00000540567.1 |
ATG16L2
|
autophagy related 16 like 2 |
| chr16_+_33009175 | 2.65 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr17_-_19046957 | 2.64 |
ENST00000284154.10
ENST00000573099.5 |
GRAP
|
GRB2 related adaptor protein |
| chr14_-_106470788 | 2.60 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr14_+_103123452 | 2.59 |
ENST00000558056.1
ENST00000560869.6 |
TNFAIP2
|
TNF alpha induced protein 2 |
| chr5_+_136058849 | 2.56 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced |
| chr19_+_14031746 | 2.53 |
ENST00000263379.4
|
IL27RA
|
interleukin 27 receptor subunit alpha |
| chr5_-_150412743 | 2.52 |
ENST00000353334.11
ENST00000009530.12 ENST00000377795.7 |
CD74
|
CD74 molecule |
| chr12_+_6946468 | 2.51 |
ENST00000543115.5
ENST00000399448.5 |
PTPN6
|
protein tyrosine phosphatase non-receptor type 6 |
| chr16_+_32995048 | 2.49 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr13_-_46182136 | 2.48 |
ENST00000323076.7
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr14_+_51847145 | 2.45 |
ENST00000615906.4
|
GNG2
|
G protein subunit gamma 2 |
| chr7_+_150800403 | 2.45 |
ENST00000484928.5
|
TMEM176A
|
transmembrane protein 176A |
| chr16_+_72054477 | 2.42 |
ENST00000355906.10
ENST00000570083.5 ENST00000228226.12 ENST00000398131.6 ENST00000569639.5 ENST00000564499.5 ENST00000357763.8 ENST00000613898.1 ENST00000562526.5 ENST00000565574.5 ENST00000568417.6 |
HP
|
haptoglobin |
| chr9_+_124291935 | 2.40 |
ENST00000546191.5
|
NEK6
|
NIMA related kinase 6 |
| chr2_+_218880844 | 2.39 |
ENST00000258411.8
|
WNT10A
|
Wnt family member 10A |
| chr3_-_58210961 | 2.39 |
ENST00000486455.5
ENST00000394549.7 |
DNASE1L3
|
deoxyribonuclease 1 like 3 |
| chr6_-_32941018 | 2.37 |
ENST00000418107.3
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr11_-_104968533 | 2.37 |
ENST00000444739.7
|
CASP4
|
caspase 4 |
| chr6_+_29942523 | 2.31 |
ENST00000376809.10
ENST00000376802.2 ENST00000638375.1 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr14_-_106389858 | 2.28 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr15_-_78944985 | 2.23 |
ENST00000615999.5
ENST00000677789.1 ENST00000676880.1 ENST00000677936.1 ENST00000220166.10 ENST00000677810.1 ENST00000678644.1 ENST00000677534.1 ENST00000677316.1 |
CTSH
|
cathepsin H |
| chr6_-_32668368 | 2.20 |
ENST00000399084.5
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr11_+_118883884 | 2.20 |
ENST00000292174.5
|
CXCR5
|
C-X-C motif chemokine receptor 5 |
| chr11_+_18266254 | 2.18 |
ENST00000532858.5
ENST00000649195.1 ENST00000356524.9 ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr6_-_73520985 | 2.17 |
ENST00000676710.1
ENST00000316292.13 ENST00000309268.11 ENST00000610520.5 ENST00000678515.1 ENST00000678703.1 |
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr17_+_19127535 | 2.15 |
ENST00000577213.1
ENST00000344415.9 |
GRAPL
|
GRB2 related adaptor protein like |
| chr14_+_75522531 | 2.10 |
ENST00000555504.1
|
BATF
|
basic leucine zipper ATF-like transcription factor |
| chr4_-_163332589 | 2.10 |
ENST00000296533.3
ENST00000509586.5 ENST00000504391.5 ENST00000512819.1 |
NPY1R
|
neuropeptide Y receptor Y1 |
| chr2_+_201183120 | 2.09 |
ENST00000272879.9
ENST00000286186.11 ENST00000374650.7 ENST00000346817.9 ENST00000313728.11 ENST00000448480.1 |
CASP10
|
caspase 10 |
| chr7_+_99374240 | 2.08 |
ENST00000443222.6
ENST00000414376.5 |
ARPC1B
|
actin related protein 2/3 complex subunit 1B |
| chr17_-_28549333 | 2.06 |
ENST00000470125.5
|
UNC119
|
unc-119 lipid binding chaperone |
| chr14_+_22304051 | 2.05 |
ENST00000390466.1
|
TRAV39
|
T cell receptor alpha variable 39 |
| chr12_-_54973683 | 2.04 |
ENST00000532804.5
ENST00000531122.5 ENST00000533446.5 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr10_+_30434021 | 2.03 |
ENST00000542547.5
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr6_+_31587049 | 2.01 |
ENST00000376089.6
ENST00000396112.6 |
LST1
|
leukocyte specific transcript 1 |
| chr11_-_75351686 | 1.99 |
ENST00000360025.7
|
ARRB1
|
arrestin beta 1 |
| chr3_-_58211212 | 1.99 |
ENST00000461914.7
|
DNASE1L3
|
deoxyribonuclease 1 like 3 |
| chr6_-_44265541 | 1.97 |
ENST00000619360.6
|
NFKBIE
|
NFKB inhibitor epsilon |
| chrX_-_30577759 | 1.96 |
ENST00000378962.4
|
TASL
|
TLR adaptor interacting with endolysosomal SLC15A4 |
| chr4_-_108166289 | 1.95 |
ENST00000512172.1
|
LEF1
|
lymphoid enhancer binding factor 1 |
| chr4_+_154563003 | 1.95 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr17_+_7023042 | 1.93 |
ENST00000293805.10
|
BCL6B
|
BCL6B transcription repressor |
| chr9_+_90801757 | 1.92 |
ENST00000375751.8
ENST00000375754.9 |
SYK
|
spleen associated tyrosine kinase |
| chr1_-_169630115 | 1.92 |
ENST00000263686.11
ENST00000367788.6 |
SELP
|
selectin P |
| chr6_-_73521549 | 1.91 |
ENST00000676547.1
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr19_+_45001430 | 1.89 |
ENST00000625761.2
ENST00000505236.1 ENST00000221452.13 |
RELB
|
RELB proto-oncogene, NF-kB subunit |
| chr15_+_90184912 | 1.89 |
ENST00000561085.1
ENST00000332496.10 |
SEMA4B
|
semaphorin 4B |
| chr16_+_57358775 | 1.85 |
ENST00000219235.5
|
CCL22
|
C-C motif chemokine ligand 22 |
| chr22_+_38957522 | 1.85 |
ENST00000618553.1
ENST00000249116.7 |
APOBEC3A
|
apolipoprotein B mRNA editing enzyme catalytic subunit 3A |
| chr6_-_31736504 | 1.84 |
ENST00000616760.1
ENST00000375784.7 ENST00000375779.6 |
CLIC1
|
chloride intracellular channel 1 |
| chr12_+_5432101 | 1.78 |
ENST00000423158.4
|
NTF3
|
neurotrophin 3 |
| chr11_-_75351609 | 1.78 |
ENST00000420843.7
|
ARRB1
|
arrestin beta 1 |
| chr2_-_74553934 | 1.77 |
ENST00000264094.8
ENST00000393937.6 ENST00000409986.5 |
LOXL3
|
lysyl oxidase like 3 |
| chr1_-_169630076 | 1.74 |
ENST00000367786.6
ENST00000458599.6 ENST00000367795.2 |
SELP
|
selectin P |
| chr16_+_3065297 | 1.73 |
ENST00000325568.9
|
IL32
|
interleukin 32 |
| chr9_-_128724088 | 1.72 |
ENST00000406904.2
ENST00000452105.5 ENST00000372667.9 ENST00000372663.9 |
ZDHHC12
|
zinc finger DHHC-type palmitoyltransferase 12 |
| chr19_-_6591103 | 1.70 |
ENST00000423145.7
ENST00000245903.4 |
CD70
|
CD70 molecule |
| chr19_-_51630401 | 1.69 |
ENST00000683636.1
|
SIGLEC5
|
sialic acid binding Ig like lectin 5 |
| chr14_+_21965451 | 1.69 |
ENST00000390442.3
|
TRAV12-3
|
T cell receptor alpha variable 12-3 |
| chr12_-_57478042 | 1.69 |
ENST00000548139.5
|
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr4_+_85827891 | 1.69 |
ENST00000514229.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr7_-_22220226 | 1.66 |
ENST00000420196.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr6_-_73521783 | 1.64 |
ENST00000331523.7
ENST00000356303.7 |
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr16_+_56936654 | 1.61 |
ENST00000563911.5
|
HERPUD1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr9_+_90801909 | 1.60 |
ENST00000375747.5
|
SYK
|
spleen associated tyrosine kinase |
| chr7_-_100827504 | 1.59 |
ENST00000616502.4
ENST00000358173.8 |
EPHB4
|
EPH receptor B4 |
| chr2_+_236569817 | 1.59 |
ENST00000272928.4
|
ACKR3
|
atypical chemokine receptor 3 |
| chr2_+_73892967 | 1.58 |
ENST00000409731.7
ENST00000409918.5 ENST00000442912.5 ENST00000345517.8 ENST00000409624.1 |
ACTG2
|
actin gamma 2, smooth muscle |
| chr10_-_71773513 | 1.57 |
ENST00000394957.8
|
VSIR
|
V-set immunoregulatory receptor |
| chr11_-_105023136 | 1.55 |
ENST00000526056.5
ENST00000531367.5 ENST00000456094.1 ENST00000444749.6 ENST00000393141.6 ENST00000418434.5 ENST00000260315.8 |
CASP5
|
caspase 5 |
| chr10_+_30434176 | 1.54 |
ENST00000263056.6
ENST00000375322.2 |
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr1_+_23791962 | 1.53 |
ENST00000421070.1
|
LYPLA2
|
lysophospholipase 2 |
| chr20_+_45891348 | 1.52 |
ENST00000419493.3
|
CTSA
|
cathepsin A |
| chrX_-_101407893 | 1.50 |
ENST00000676156.1
ENST00000675592.1 ENST00000674634.2 ENST00000649178.1 ENST00000218516.4 |
GLA
|
galactosidase alpha |
| chr4_+_85827745 | 1.50 |
ENST00000509300.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr1_-_47231715 | 1.49 |
ENST00000371884.6
|
TAL1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr17_-_64020566 | 1.47 |
ENST00000578313.5
ENST00000584084.1 ENST00000579687.5 ENST00000578379.5 ENST00000578892.5 ENST00000579788.6 ENST00000412356.5 ENST00000418105.5 |
ICAM2
|
intercellular adhesion molecule 2 |
| chr17_-_1486124 | 1.46 |
ENST00000575158.5
|
MYO1C
|
myosin IC |
| chr11_-_57426745 | 1.45 |
ENST00000529554.5
|
SLC43A3
|
solute carrier family 43 member 3 |
| chr4_+_39044995 | 1.44 |
ENST00000261425.7
ENST00000508137.6 |
KLHL5
|
kelch like family member 5 |
| chr2_-_43226594 | 1.44 |
ENST00000282388.4
|
ZFP36L2
|
ZFP36 ring finger protein like 2 |
| chr10_+_30434116 | 1.43 |
ENST00000415139.5
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr6_+_33391805 | 1.43 |
ENST00000428849.7
ENST00000450504.1 |
KIFC1
|
kinesin family member C1 |
| chr6_+_137867241 | 1.42 |
ENST00000612899.5
ENST00000420009.5 |
TNFAIP3
|
TNF alpha induced protein 3 |
| chr6_+_32637419 | 1.41 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr2_+_96816236 | 1.41 |
ENST00000377060.7
ENST00000305510.4 |
CNNM3
|
cyclin and CBS domain divalent metal cation transport mediator 3 |
| chr1_+_111227699 | 1.40 |
ENST00000369748.9
|
CHI3L2
|
chitinase 3 like 2 |
| chr16_-_11976611 | 1.40 |
ENST00000538896.5
ENST00000673243.1 |
NPIPB2
|
nuclear pore complex interacting protein family member B2 |
| chr17_-_45317006 | 1.40 |
ENST00000344686.8
ENST00000617331.3 |
MAP3K14
|
mitogen-activated protein kinase kinase kinase 14 |
| chr17_-_1485733 | 1.39 |
ENST00000648446.1
|
MYO1C
|
myosin IC |
| chr6_+_31587002 | 1.39 |
ENST00000376090.6
|
LST1
|
leukocyte specific transcript 1 |
| chr5_+_50666612 | 1.39 |
ENST00000281631.10
|
PARP8
|
poly(ADP-ribose) polymerase family member 8 |
| chr5_-_39424966 | 1.38 |
ENST00000515700.5
ENST00000320816.11 ENST00000339788.10 |
DAB2
|
DAB adaptor protein 2 |
| chr19_-_41363930 | 1.38 |
ENST00000675972.1
|
B9D2
|
B9 domain containing 2 |
| chr19_+_35000275 | 1.37 |
ENST00000317991.10
ENST00000680623.1 |
GRAMD1A
|
GRAM domain containing 1A |
| chr12_-_108598036 | 1.37 |
ENST00000392806.4
ENST00000547567.1 |
TMEM119
|
transmembrane protein 119 |
| chr8_+_24384275 | 1.36 |
ENST00000256412.8
|
ADAMDEC1
|
ADAM like decysin 1 |
| chr14_+_22320128 | 1.36 |
ENST00000390468.1
|
TRAV41
|
T cell receptor alpha variable 41 |
| chr17_+_42288076 | 1.36 |
ENST00000546010.6
|
STAT5A
|
signal transducer and activator of transcription 5A |
| chr16_-_11587450 | 1.35 |
ENST00000571688.5
|
LITAF
|
lipopolysaccharide induced TNF factor |
| chr12_-_47905003 | 1.35 |
ENST00000550325.5
ENST00000546653.5 ENST00000548664.1 |
VDR
|
vitamin D receptor |
| chr7_-_98252117 | 1.35 |
ENST00000420697.1
ENST00000415086.5 ENST00000447648.7 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr1_+_111227610 | 1.35 |
ENST00000369744.6
|
CHI3L2
|
chitinase 3 like 2 |
| chr19_+_41363989 | 1.33 |
ENST00000413014.6
|
TMEM91
|
transmembrane protein 91 |
| chr9_+_129665603 | 1.32 |
ENST00000372469.6
|
PRRX2
|
paired related homeobox 2 |
| chr20_+_45891309 | 1.32 |
ENST00000354880.9
ENST00000646241.3 ENST00000191018.9 |
CTSA
|
cathepsin A |
| chr8_+_27311620 | 1.32 |
ENST00000522338.5
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr12_-_53200443 | 1.31 |
ENST00000550743.6
|
ITGB7
|
integrin subunit beta 7 |
| chr15_+_49423233 | 1.29 |
ENST00000560270.1
ENST00000267843.9 ENST00000560979.1 |
FGF7
|
fibroblast growth factor 7 |
| chr14_+_51847116 | 1.29 |
ENST00000553560.5
|
GNG2
|
G protein subunit gamma 2 |
| chr5_+_50666660 | 1.28 |
ENST00000515175.6
|
PARP8
|
poly(ADP-ribose) polymerase family member 8 |
| chr12_-_47904986 | 1.28 |
ENST00000549336.6
|
VDR
|
vitamin D receptor |
| chr22_+_50481515 | 1.27 |
ENST00000395737.2
ENST00000395738.2 |
ADM2
|
adrenomedullin 2 |
| chr14_-_95516616 | 1.26 |
ENST00000682763.1
|
SYNE3
|
spectrin repeat containing nuclear envelope family member 3 |
| chr22_+_39960397 | 1.26 |
ENST00000424496.2
|
ENSG00000225528.3
|
novel protein similar to translation machinery associated 7 homolog (S. cerevisiae) TMA7 |
| chr17_+_7023062 | 1.26 |
ENST00000576705.1
|
BCL6B
|
BCL6B transcription repressor |
| chr9_-_120929160 | 1.25 |
ENST00000540010.1
|
TRAF1
|
TNF receptor associated factor 1 |
| chr5_-_39424859 | 1.23 |
ENST00000503513.5
|
DAB2
|
DAB adaptor protein 2 |
| chr14_+_22086401 | 1.23 |
ENST00000390451.2
|
TRAV23DV6
|
T cell receptor alpha variable 23/delta variable 6 |
| chr1_-_151993822 | 1.21 |
ENST00000368811.8
|
S100A10
|
S100 calcium binding protein A10 |
| chr19_+_34994778 | 1.21 |
ENST00000599564.5
|
GRAMD1A
|
GRAM domain containing 1A |
| chr6_+_32637396 | 1.21 |
ENST00000395363.5
ENST00000496318.5 ENST00000343139.11 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr2_+_20667136 | 1.21 |
ENST00000272224.5
|
GDF7
|
growth differentiation factor 7 |
| chr19_+_926001 | 1.21 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr20_-_47785439 | 1.21 |
ENST00000437955.1
|
SULF2
|
sulfatase 2 |
| chr11_+_59142811 | 1.21 |
ENST00000676459.1
ENST00000675163.1 ENST00000684135.1 ENST00000682018.1 ENST00000675806.2 ENST00000529985.3 ENST00000676340.1 ENST00000674617.1 |
FAM111A
|
FAM111 trypsin like peptidase A |
| chr19_-_55258942 | 1.21 |
ENST00000412770.7
|
PPP6R1
|
protein phosphatase 6 regulatory subunit 1 |
| chr8_+_24384455 | 1.20 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM like decysin 1 |
| chr1_+_113979460 | 1.19 |
ENST00000320334.5
|
OLFML3
|
olfactomedin like 3 |
| chr1_-_151992571 | 1.18 |
ENST00000368809.1
|
S100A10
|
S100 calcium binding protein A10 |
| chr4_-_76023489 | 1.17 |
ENST00000306602.3
|
CXCL10
|
C-X-C motif chemokine ligand 10 |
| chr6_+_137867414 | 1.16 |
ENST00000237289.8
ENST00000433680.1 |
TNFAIP3
|
TNF alpha induced protein 3 |
| chr12_+_112013348 | 1.16 |
ENST00000455836.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr20_+_63735463 | 1.16 |
ENST00000496820.2
|
ENSG00000273154.3
|
novel protein, ZGPAT-LIME1 readthrough |
| chr15_+_90868580 | 1.16 |
ENST00000268171.8
|
FURIN
|
furin, paired basic amino acid cleaving enzyme |
| chr19_+_40577158 | 1.15 |
ENST00000595631.5
|
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr1_-_159924529 | 1.15 |
ENST00000320307.8
|
TAGLN2
|
transgelin 2 |
| chr4_-_80073170 | 1.15 |
ENST00000403729.7
|
ANTXR2
|
ANTXR cell adhesion molecule 2 |
| chr17_-_55421818 | 1.15 |
ENST00000262065.8
ENST00000649377.1 |
MMD
|
monocyte to macrophage differentiation associated |
| chr5_-_139482685 | 1.14 |
ENST00000651565.1
|
STING1
|
stimulator of interferon response cGAMP interactor 1 |
| chr12_+_57230086 | 1.13 |
ENST00000414700.7
ENST00000557703.5 |
SHMT2
|
serine hydroxymethyltransferase 2 |
| chr11_-_57426638 | 1.13 |
ENST00000529112.5
ENST00000529896.1 |
SLC43A3
|
solute carrier family 43 member 3 |
| chr6_-_27472681 | 1.11 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr12_-_57110284 | 1.11 |
ENST00000543873.6
ENST00000554663.5 ENST00000557635.5 |
STAT6
|
signal transducer and activator of transcription 6 |
| chr4_-_80072993 | 1.11 |
ENST00000681115.1
|
ANTXR2
|
ANTXR cell adhesion molecule 2 |
| chr12_+_112013418 | 1.11 |
ENST00000261735.4
ENST00000552052.1 |
ERP29
|
endoplasmic reticulum protein 29 |
| chr13_+_30713477 | 1.11 |
ENST00000617770.4
|
ALOX5AP
|
arachidonate 5-lipoxygenase activating protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of HIVEP1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 1.5 | 15.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 1.2 | 8.7 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 1.0 | 3.1 | GO:2000627 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.9 | 2.8 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.9 | 3.5 | GO:0002238 | response to molecule of fungal origin(GO:0002238) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) cellular response to molecule of fungal origin(GO:0071226) |
| 0.8 | 7.6 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.8 | 2.5 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.8 | 71.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.8 | 2.4 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.8 | 10.3 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.7 | 4.1 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.7 | 2.8 | GO:0034148 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.7 | 2.1 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.6 | 6.7 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.6 | 0.6 | GO:2000426 | negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.6 | 5.3 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.6 | 2.3 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.6 | 2.2 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.6 | 1.7 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.5 | 1.1 | GO:0090291 | negative regulation of osteoclast proliferation(GO:0090291) |
| 0.5 | 2.6 | GO:0060058 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.5 | 0.5 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.5 | 2.6 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.5 | 3.0 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.5 | 2.5 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.5 | 1.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.5 | 3.0 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.5 | 1.5 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.5 | 1.5 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.5 | 2.7 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.4 | 1.3 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.4 | 45.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.4 | 3.8 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.4 | 0.7 | GO:0046645 | positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.4 | 3.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.4 | 1.1 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.4 | 1.8 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.3 | 1.0 | GO:1990051 | activation of protein kinase C activity(GO:1990051) regulation of glutamine transport(GO:2000485) |
| 0.3 | 0.7 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.3 | 1.0 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.3 | 3.2 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.3 | 1.0 | GO:0000494 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.3 | 8.6 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.3 | 4.0 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.3 | 1.8 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.3 | 1.2 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.3 | 3.2 | GO:1901164 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) negative regulation of trophoblast cell migration(GO:1901164) |
| 0.3 | 5.2 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.3 | 3.5 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.3 | 1.3 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.3 | 1.6 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.2 | 0.7 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.2 | 5.5 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.2 | 2.4 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 | 1.9 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.2 | 2.5 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.2 | 1.1 | GO:0043373 | T-helper cell lineage commitment(GO:0002295) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) |
| 0.2 | 1.5 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.2 | 0.7 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.2 | 4.0 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.2 | 1.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.2 | 2.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.2 | 3.7 | GO:0033623 | positive regulation of platelet activation(GO:0010572) regulation of integrin activation(GO:0033623) |
| 0.2 | 2.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.2 | 3.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 3.0 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 5.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 0.5 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 3.5 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.2 | 2.2 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.2 | 1.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 1.0 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 1.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 0.9 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.2 | 1.5 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.2 | 2.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 1.2 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 0.8 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.2 | 1.5 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 2.4 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 1.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.8 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 2.2 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) |
| 0.1 | 3.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.9 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 2.3 | GO:0032329 | serine transport(GO:0032329) |
| 0.1 | 1.0 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 | 0.6 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.1 | 0.4 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) histone H3-K9 deacetylation(GO:1990619) |
| 0.1 | 1.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.8 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 1.0 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.8 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 2.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.1 | 0.4 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.1 | 13.6 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 1.0 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 3.7 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.5 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.1 | 2.4 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.1 | 11.9 | GO:0032945 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.1 | 0.4 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.1 | 1.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.8 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 3.1 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 1.6 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.2 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.1 | 0.7 | GO:0046641 | positive regulation of alpha-beta T cell proliferation(GO:0046641) |
| 0.1 | 3.4 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.1 | 0.3 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 0.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.8 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 1.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 2.2 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.1 | 1.0 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.3 | GO:2000910 | negative regulation of triglyceride catabolic process(GO:0010897) negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.1 | 1.5 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 0.2 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.1 | 0.2 | GO:1902960 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 2.7 | GO:0042092 | type 2 immune response(GO:0042092) |
| 0.1 | 0.3 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.1 | 0.6 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 2.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.5 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.6 | GO:0046070 | dGTP metabolic process(GO:0046070) |
| 0.1 | 2.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.9 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 0.4 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.4 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 1.7 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 0.8 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.2 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) negative regulation of transforming growth factor-beta secretion(GO:2001202) |
| 0.1 | 2.5 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 1.0 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 1.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.4 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 5.0 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.7 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 2.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.5 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.2 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 1.4 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.3 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.3 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.3 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.3 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.5 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 1.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.9 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 2.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.8 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 1.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.8 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.8 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 2.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.1 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.0 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 3.3 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.1 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.3 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.0 | 1.9 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.6 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 1.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 1.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 1.5 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.2 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.3 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.4 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 1.1 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.7 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 1.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.4 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 1.8 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 1.0 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 1.3 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.0 | 0.6 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 1.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.6 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.0 | 1.8 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 3.1 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.0 | 1.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 1.2 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 2.9 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 1.1 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 4.1 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 0.3 | GO:0032625 | interleukin-21 production(GO:0032625) chemokine (C-C motif) ligand 2 secretion(GO:0035926) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 1.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 | 0.8 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.4 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 1.1 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.6 | GO:0046632 | alpha-beta T cell differentiation(GO:0046632) |
| 0.0 | 1.5 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.6 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.2 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.3 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 7.8 | GO:0071757 | hexameric IgM immunoglobulin complex(GO:0071757) |
| 1.4 | 15.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.2 | 63.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.9 | 17.6 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.7 | 3.5 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.7 | 2.1 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.7 | 3.9 | GO:0097169 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 0.6 | 2.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.6 | 2.9 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.4 | 8.7 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.4 | 5.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.4 | 5.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.4 | 1.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 3.7 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.3 | 2.6 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 2.8 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.3 | 3.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 3.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.3 | 1.3 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.2 | 2.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 1.6 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 1.0 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.2 | 0.8 | GO:0043293 | apoptosome(GO:0043293) |
| 0.2 | 1.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 4.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 2.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.2 | 2.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 1.0 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 3.7 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 3.9 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 1.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.5 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 0.3 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 3.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 1.4 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 3.0 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 1.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 4.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 2.2 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 1.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 2.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.5 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 2.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 2.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.4 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 7.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 4.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 12.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 2.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.7 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 5.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 2.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 2.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 1.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 19.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 1.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 1.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 2.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 3.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 1.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.7 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 6.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 2.5 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 3.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 2.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 1.0 | 71.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.9 | 2.6 | GO:1902098 | calcitriol receptor activity(GO:0008434) lithocholic acid receptor activity(GO:0038186) calcitriol binding(GO:1902098) lithocholic acid binding(GO:1902121) |
| 0.8 | 4.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.8 | 6.5 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.7 | 12.7 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.6 | 5.3 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.5 | 8.7 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.5 | 2.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.5 | 3.7 | GO:0042806 | fucose binding(GO:0042806) |
| 0.5 | 2.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.4 | 3.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.4 | 6.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.4 | 1.9 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.4 | 1.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.3 | 1.0 | GO:0009032 | thymidine phosphorylase activity(GO:0009032) pyrimidine-nucleoside phosphorylase activity(GO:0016154) |
| 0.3 | 2.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 1.0 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.3 | 63.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.3 | 1.0 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.3 | 3.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 2.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 2.5 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.2 | 1.0 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.2 | 2.8 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.2 | 3.8 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 1.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.2 | 1.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 3.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 5.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 3.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 3.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 0.5 | GO:0032428 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.2 | 1.8 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 3.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.2 | 5.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 0.5 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.2 | 3.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 3.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 1.0 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 1.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.2 | 5.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 0.8 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 0.6 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 2.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 3.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 1.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 2.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 2.9 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.1 | 0.9 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.4 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 3.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 3.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.6 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.1 | 2.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 5.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.5 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.4 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 0.8 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.4 | GO:0046921 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 0.7 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 4.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.9 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.5 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.1 | 0.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.4 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.1 | 2.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 1.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 3.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 1.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 2.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.3 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.1 | 0.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.6 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.3 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 0.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 4.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.6 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 2.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 3.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.3 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.4 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 2.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 2.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.6 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 1.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.9 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 2.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 3.7 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 1.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.3 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 2.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 5.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.9 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 1.0 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 2.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.4 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 1.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.0 | 1.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 1.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 5.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 6.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 6.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 14.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 10.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 10.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 5.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 9.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 3.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 3.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 4.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 0.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 1.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 4.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 0.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 4.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 5.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 2.0 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 2.9 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 9.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 4.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 2.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 3.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 2.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 3.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 2.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 7.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 3.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 4.4 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 15.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.5 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 6.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.3 | 2.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.2 | 7.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 2.9 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 8.6 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.2 | 8.6 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 11.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.2 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.1 | 1.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 6.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 4.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 7.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 4.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 3.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 8.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 3.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 4.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 8.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 5.0 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 2.2 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.6 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 1.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 5.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 3.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 2.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 2.4 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 3.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |