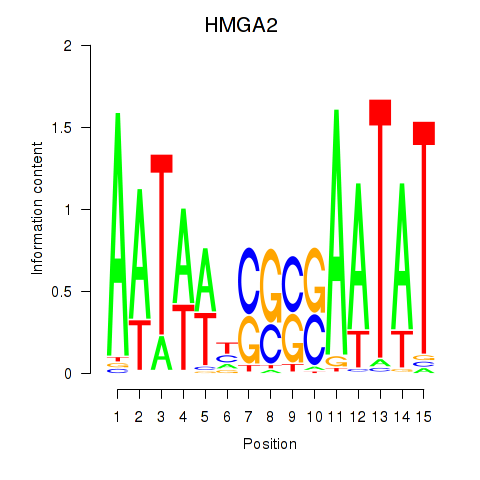
|
chr2_+_1413456
Show fit
|
3.84 |
ENST00000539820.5
ENST00000382269.7
ENST00000345913.8
ENST00000329066.9
ENST00000382201.7
|
TPO
|
thyroid peroxidase
|
|
chr11_-_19201976
Show fit
|
3.76 |
ENST00000647990.1
ENST00000649235.1
ENST00000265968.9
ENST00000649842.1
|
CSRP3
|
cysteine and glycine rich protein 3
|
|
chr15_-_53759634
Show fit
|
2.67 |
ENST00000557913.5
ENST00000360509.10
|
WDR72
|
WD repeat domain 72
|
|
chr10_-_5003850
Show fit
|
2.21 |
ENST00000421196.7
ENST00000455190.2
ENST00000380753.8
|
AKR1C2
|
aldo-keto reductase family 1 member C2
|
|
chr11_-_19202004
Show fit
|
1.96 |
ENST00000648719.1
|
CSRP3
|
cysteine and glycine rich protein 3
|
|
chr8_-_104467042
Show fit
|
1.92 |
ENST00000521573.2
ENST00000351513.7
|
DPYS
|
dihydropyrimidinase
|
|
chr8_+_119207949
Show fit
|
1.72 |
ENST00000534619.5
|
MAL2
|
mal, T cell differentiation protein 2
|
|
chr15_+_58410543
Show fit
|
1.59 |
ENST00000356113.10
ENST00000414170.7
|
LIPC
|
lipase C, hepatic type
|
|
chr10_-_127892930
Show fit
|
1.45 |
ENST00000368671.4
|
CLRN3
|
clarin 3
|
|
chr22_-_18936142
Show fit
|
1.45 |
ENST00000438924.5
ENST00000457083.1
ENST00000357068.11
ENST00000420436.5
ENST00000334029.6
ENST00000610940.4
|
PRODH
|
proline dehydrogenase 1
|
|
chr14_-_94293071
Show fit
|
1.27 |
ENST00000554723.5
|
SERPINA10
|
serpin family A member 10
|
|
chr12_+_18261511
Show fit
|
1.26 |
ENST00000538779.6
ENST00000433979.6
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma
|
|
chr14_-_94293024
Show fit
|
1.25 |
ENST00000393096.5
|
SERPINA10
|
serpin family A member 10
|
|
chr1_-_75932392
Show fit
|
1.17 |
ENST00000284142.7
|
ASB17
|
ankyrin repeat and SOCS box containing 17
|
|
chr1_+_196977550
Show fit
|
1.09 |
ENST00000256785.5
|
CFHR5
|
complement factor H related 5
|
|
chrX_+_2752024
Show fit
|
1.08 |
ENST00000644266.2
ENST00000419513.7
ENST00000509484.3
ENST00000381174.10
|
XG
|
Xg glycoprotein (Xg blood group)
|
|
chr14_-_94293260
Show fit
|
1.04 |
ENST00000261994.9
|
SERPINA10
|
serpin family A member 10
|
|
chr1_+_196652022
Show fit
|
0.89 |
ENST00000367429.9
ENST00000630130.2
ENST00000359637.2
|
CFH
|
complement factor H
|
|
chr3_+_155083523
Show fit
|
0.87 |
ENST00000680057.1
|
MME
|
membrane metalloendopeptidase
|
|
chr9_+_105700953
Show fit
|
0.85 |
ENST00000374688.5
|
TMEM38B
|
transmembrane protein 38B
|
|
chrX_-_7927701
Show fit
|
0.74 |
ENST00000537427.5
ENST00000444736.5
ENST00000442940.1
|
PNPLA4
|
patatin like phospholipase domain containing 4
|
|
chr9_-_24545866
Show fit
|
0.74 |
ENST00000543880.7
ENST00000418122.1
|
IZUMO3
|
IZUMO family member 3
|
|
chr7_+_80638510
Show fit
|
0.72 |
ENST00000433696.6
ENST00000538969.5
ENST00000544133.5
|
CD36
|
CD36 molecule
|
|
chr18_+_11851404
Show fit
|
0.71 |
ENST00000526991.3
|
CHMP1B
|
charged multivesicular body protein 1B
|
|
chr16_-_20404731
Show fit
|
0.71 |
ENST00000302451.9
|
PDILT
|
protein disulfide isomerase like, testis expressed
|
|
chr2_+_119679154
Show fit
|
0.70 |
ENST00000401466.5
ENST00000424086.5
|
TMEM177
|
transmembrane protein 177
|
|
chrX_+_23908006
Show fit
|
0.67 |
ENST00000379211.8
ENST00000648352.1
|
CXorf58
|
chromosome X open reading frame 58
|
|
chr2_+_234978685
Show fit
|
0.65 |
ENST00000409212.5
ENST00000344528.8
ENST00000444916.5
|
SH3BP4
|
SH3 domain binding protein 4
|
|
chr9_-_34729482
Show fit
|
0.64 |
ENST00000378788.4
|
FAM205A
|
family with sequence similarity 205 member A
|
|
chr19_-_11346406
Show fit
|
0.62 |
ENST00000587948.5
|
TMEM205
|
transmembrane protein 205
|
|
chrX_-_7927375
Show fit
|
0.60 |
ENST00000381042.9
|
PNPLA4
|
patatin like phospholipase domain containing 4
|
|
chr2_+_169827432
Show fit
|
0.60 |
ENST00000272793.11
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3
|
|
chr15_-_74367637
Show fit
|
0.59 |
ENST00000268053.11
ENST00000416978.1
|
CYP11A1
|
cytochrome P450 family 11 subfamily A member 1
|
|
chr6_-_49866453
Show fit
|
0.58 |
ENST00000507853.5
|
CRISP1
|
cysteine rich secretory protein 1
|
|
chr2_+_119679184
Show fit
|
0.57 |
ENST00000445518.1
ENST00000272521.7
ENST00000409951.1
|
TMEM177
|
transmembrane protein 177
|
|
chr11_-_130916437
Show fit
|
0.57 |
ENST00000533214.1
ENST00000528555.5
ENST00000530356.5
ENST00000265909.9
|
SNX19
|
sorting nexin 19
|
|
chr15_-_32403195
Show fit
|
0.56 |
ENST00000512626.2
|
GOLGA8K
|
golgin A8 family member K
|
|
chr1_+_103561757
Show fit
|
0.55 |
ENST00000435302.5
|
AMY2B
|
amylase alpha 2B
|
|
chr14_-_64942783
Show fit
|
0.54 |
ENST00000612794.1
|
GPX2
|
glutathione peroxidase 2
|
|
chr15_+_23354846
Show fit
|
0.53 |
ENST00000562295.1
|
GOLGA8S
|
golgin A8 family member S
|
|
chr11_+_60327250
Show fit
|
0.53 |
ENST00000684409.1
|
MS4A6E
|
membrane spanning 4-domains A6E
|
|
chr19_+_41376692
Show fit
|
0.52 |
ENST00000447302.6
ENST00000544232.5
ENST00000542945.5
ENST00000540732.3
|
TMEM91
ENSG00000255730.5
|
transmembrane protein 91
novel protein
|
|
chr15_+_30604028
Show fit
|
0.51 |
ENST00000566740.2
|
GOLGA8H
|
golgin A8 family member H
|
|
chr4_+_40056790
Show fit
|
0.49 |
ENST00000261435.11
ENST00000515550.1
|
N4BP2
|
NEDD4 binding protein 2
|
|
chr19_-_11346486
Show fit
|
0.48 |
ENST00000590482.5
|
TMEM205
|
transmembrane protein 205
|
|
chr4_+_78551733
Show fit
|
0.48 |
ENST00000512884.5
ENST00000512542.5
ENST00000503570.6
ENST00000264908.11
|
ANXA3
|
annexin A3
|
|
chr12_+_69585434
Show fit
|
0.48 |
ENST00000299300.11
ENST00000544368.6
|
CCT2
|
chaperonin containing TCP1 subunit 2
|
|
chr6_+_111259474
Show fit
|
0.48 |
ENST00000672554.1
ENST00000673024.1
|
MFSD4B
|
major facilitator superfamily domain containing 4B
|
|
chr17_-_3398410
Show fit
|
0.47 |
ENST00000322608.2
|
OR1E1
|
olfactory receptor family 1 subfamily E member 1
|
|
chr2_-_182522703
Show fit
|
0.46 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A
|
|
chr19_-_12365628
Show fit
|
0.45 |
ENST00000438182.5
|
ZNF442
|
zinc finger protein 442
|
|
chr15_+_43593054
Show fit
|
0.44 |
ENST00000453782.5
ENST00000300283.10
ENST00000437924.5
|
CKMT1B
|
creatine kinase, mitochondrial 1B
|
|
chr19_-_12365655
Show fit
|
0.44 |
ENST00000242804.9
|
ZNF442
|
zinc finger protein 442
|
|
chr6_+_31919984
Show fit
|
0.43 |
ENST00000413154.5
|
C2
|
complement C2
|
|
chrX_-_77895546
Show fit
|
0.42 |
ENST00000358075.11
|
MAGT1
|
magnesium transporter 1
|
|
chr1_+_54998927
Show fit
|
0.42 |
ENST00000651561.1
|
BSND
|
barttin CLCNK type accessory subunit beta
|
|
chr7_+_80638662
Show fit
|
0.42 |
ENST00000394788.7
|
CD36
|
CD36 molecule
|
|
chr19_-_11346228
Show fit
|
0.40 |
ENST00000588560.5
ENST00000592952.5
|
TMEM205
|
transmembrane protein 205
|
|
chr1_-_247536440
Show fit
|
0.40 |
ENST00000366487.4
ENST00000641802.1
|
OR2C3
|
olfactory receptor family 2 subfamily C member 3
|
|
chr1_+_197917355
Show fit
|
0.39 |
ENST00000367388.4
ENST00000367387.6
|
LHX9
|
LIM homeobox 9
|
|
chr19_-_11346259
Show fit
|
0.38 |
ENST00000590788.1
ENST00000354882.10
ENST00000586590.5
ENST00000589555.5
ENST00000586956.5
ENST00000593256.6
ENST00000447337.5
ENST00000591677.5
ENST00000586701.1
ENST00000589655.1
|
TMEM205
RAB3D
|
transmembrane protein 205
RAB3D, member RAS oncogene family
|
|
chrX_+_3006546
Show fit
|
0.37 |
ENST00000381130.3
|
ARSH
|
arylsulfatase family member H
|
|
chrX_-_13319952
Show fit
|
0.36 |
ENST00000622204.1
ENST00000380622.5
|
ATXN3L
|
ataxin 3 like
|
|
chr11_+_60492775
Show fit
|
0.36 |
ENST00000526784.5
ENST00000537076.5
ENST00000016913.8
ENST00000530007.1
|
MS4A12
|
membrane spanning 4-domains A12
|
|
chr11_-_55968514
Show fit
|
0.35 |
ENST00000312345.4
|
OR10AG1
|
olfactory receptor family 10 subfamily AG member 1
|
|
chr2_+_227745845
Show fit
|
0.35 |
ENST00000641981.1
|
SCYGR8
|
small cysteine and glycine repeat containing 8
|
|
chr8_-_54101458
Show fit
|
0.34 |
ENST00000521352.5
ENST00000618741.1
|
LYPLA1
|
lysophospholipase 1
|
|
chr19_-_11346196
Show fit
|
0.34 |
ENST00000586218.5
|
TMEM205
|
transmembrane protein 205
|
|
chr6_-_127343518
Show fit
|
0.33 |
ENST00000528402.5
ENST00000454591.6
|
ECHDC1
|
ethylmalonyl-CoA decarboxylase 1
|
|
chr15_+_28378664
Show fit
|
0.33 |
ENST00000532622.7
ENST00000526619.6
|
GOLGA8F
|
golgin A8 family member F
|
|
chr15_-_32455634
Show fit
|
0.32 |
ENST00000509311.7
|
GOLGA8O
|
golgin A8 family member O
|
|
chr6_-_127343590
Show fit
|
0.32 |
ENST00000368291.6
ENST00000454859.8
|
ECHDC1
|
ethylmalonyl-CoA decarboxylase 1
|
|
chrX_-_23907887
Show fit
|
0.30 |
ENST00000379226.9
|
APOO
|
apolipoprotein O
|
|
chr14_+_88385714
Show fit
|
0.30 |
ENST00000045347.11
|
SPATA7
|
spermatogenesis associated 7
|
|
chr6_-_117573571
Show fit
|
0.29 |
ENST00000467125.1
|
ENSG00000282218.1
|
novel protein, GOPC-ROS1 readthrough
|
|
chr10_+_94545777
Show fit
|
0.29 |
ENST00000348459.10
ENST00000419900.5
ENST00000630929.2
|
HELLS
|
helicase, lymphoid specific
|
|
chr7_-_57132425
Show fit
|
0.28 |
ENST00000319636.10
|
ZNF479
|
zinc finger protein 479
|
|
chrX_-_154295085
Show fit
|
0.28 |
ENST00000617225.4
ENST00000619903.4
|
TEX28
|
testis expressed 28
|
|
chr11_-_125778788
Show fit
|
0.27 |
ENST00000436890.2
|
PATE2
|
prostate and testis expressed 2
|
|
chr11_-_125778818
Show fit
|
0.25 |
ENST00000358524.8
|
PATE2
|
prostate and testis expressed 2
|
|
chrX_-_72572794
Show fit
|
0.22 |
ENST00000421523.6
ENST00000648036.1
ENST00000373559.8
ENST00000648139.1
ENST00000650636.1
ENST00000648962.1
ENST00000373560.7
ENST00000373568.7
ENST00000373573.9
ENST00000647654.1
ENST00000647859.1
ENST00000415409.6
ENST00000648298.1
ENST00000649752.1
ENST00000373571.6
ENST00000647980.1
ENST00000650126.1
ENST00000373554.6
ENST00000648870.1
ENST00000373556.8
ENST00000647886.1
ENST00000648452.1
ENST00000647594.1
ENST00000373583.6
ENST00000648922.1
|
HDAC8
ENSG00000285547.1
|
histone deacetylase 8
novel protein
|
|
chr7_+_80638633
Show fit
|
0.22 |
ENST00000447544.7
ENST00000482059.6
|
CD36
|
CD36 molecule
|
|
chr10_-_99913971
Show fit
|
0.22 |
ENST00000543621.6
|
DNMBP
|
dynamin binding protein
|
|
chr10_-_60572599
Show fit
|
0.21 |
ENST00000503366.5
|
ANK3
|
ankyrin 3
|
|
chr8_+_66775537
Show fit
|
0.19 |
ENST00000518388.5
|
SGK3
|
serum/glucocorticoid regulated kinase family member 3
|
|
chr1_+_12746192
Show fit
|
0.18 |
ENST00000614859.5
|
C1orf158
|
chromosome 1 open reading frame 158
|
|
chr14_+_19773504
Show fit
|
0.18 |
ENST00000641200.1
|
OR4M1
|
olfactory receptor family 4 subfamily M member 1
|
|
chr4_+_183905266
Show fit
|
0.17 |
ENST00000308497.9
|
STOX2
|
storkhead box 2
|
|
chr15_-_101923113
Show fit
|
0.16 |
ENST00000650172.1
|
OR4F4
|
olfactory receptor family 4 subfamily F member 4
|
|
chrX_+_147943245
Show fit
|
0.16 |
ENST00000463120.2
|
FMR1
|
FMRP translational regulator 1
|
|
chr3_-_157503574
Show fit
|
0.15 |
ENST00000494677.5
ENST00000468233.5
|
VEPH1
|
ventricular zone expressed PH domain containing 1
|
|
chr10_+_84425148
Show fit
|
0.15 |
ENST00000493409.5
|
CCSER2
|
coiled-coil serine rich protein 2
|
|
chr5_-_55692731
Show fit
|
0.15 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38 member 9
|
|
chr15_+_84235773
Show fit
|
0.13 |
ENST00000510439.7
ENST00000422563.6
|
GOLGA6L4
|
golgin A6 family like 4
|
|
chr12_+_8157034
Show fit
|
0.12 |
ENST00000396570.7
|
ZNF705A
|
zinc finger protein 705A
|
|
chr15_-_70702273
Show fit
|
0.12 |
ENST00000558758.5
ENST00000379983.6
ENST00000560441.5
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr14_+_99481395
Show fit
|
0.11 |
ENST00000389879.9
ENST00000557441.5
ENST00000555049.5
ENST00000555842.1
|
CCNK
|
cyclin K
|
|
chr7_+_120950763
Show fit
|
0.11 |
ENST00000339121.9
ENST00000315870.10
ENST00000445699.5
|
ING3
|
inhibitor of growth family member 3
|
|
chr10_+_35167516
Show fit
|
0.10 |
ENST00000361599.8
|
CREM
|
cAMP responsive element modulator
|
|
chr2_-_182522556
Show fit
|
0.10 |
ENST00000435564.5
|
PDE1A
|
phosphodiesterase 1A
|
|
chr1_-_21113129
Show fit
|
0.10 |
ENST00000684485.1
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma 3
|
|
chr4_-_305447
Show fit
|
0.09 |
ENST00000419098.6
|
ZNF732
|
zinc finger protein 732
|
|
chr19_-_1650667
Show fit
|
0.09 |
ENST00000587235.6
|
TCF3
|
transcription factor 3
|
|
chr8_+_33485173
Show fit
|
0.08 |
ENST00000360128.11
|
MAK16
|
MAK16 homolog
|
|
chr5_-_103120097
Show fit
|
0.07 |
ENST00000508629.5
ENST00000399004.7
|
GIN1
|
gypsy retrotransposon integrase 1
|
|
chr14_-_20641691
Show fit
|
0.06 |
ENST00000320704.3
|
OR6S1
|
olfactory receptor family 6 subfamily S member 1
|
|
chr7_+_64207090
Show fit
|
0.06 |
ENST00000429565.4
|
ZNF735
|
zinc finger protein 735
|
|
chr1_+_28510050
Show fit
|
0.06 |
ENST00000419074.5
|
RCC1
|
regulator of chromosome condensation 1
|
|
chr15_-_82349437
Show fit
|
0.06 |
ENST00000621197.4
ENST00000610657.2
ENST00000619556.4
|
GOLGA6L10
|
golgin A6 family like 10
|
|
chr2_+_38889868
Show fit
|
0.05 |
ENST00000409978.7
|
ARHGEF33
|
Rho guanine nucleotide exchange factor 33
|
|
chr3_+_160842143
Show fit
|
0.05 |
ENST00000464260.5
ENST00000295839.9
|
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent 1L
|
|
chr6_-_159788439
Show fit
|
0.03 |
ENST00000539948.5
|
TCP1
|
t-complex 1
|
|
chr9_+_2157647
Show fit
|
0.02 |
ENST00000452193.5
ENST00000324954.10
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chr19_-_42594957
Show fit
|
0.01 |
ENST00000599005.1
|
CEACAM8
|
CEA cell adhesion molecule 8
|
|
chr2_-_287299
Show fit
|
0.01 |
ENST00000405290.5
|
ALKAL2
|
ALK and LTK ligand 2
|