Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
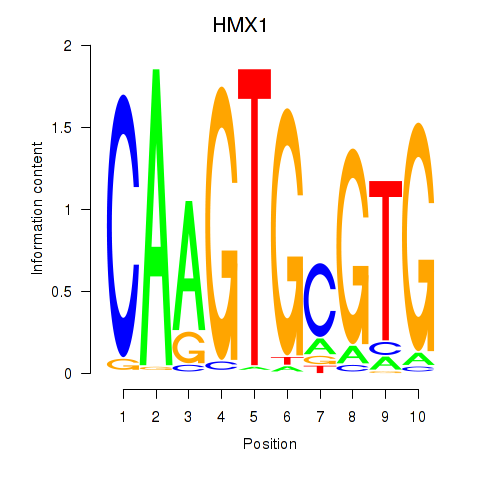
Results for HMX1
Z-value: 1.68
Transcription factors associated with HMX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX1
|
ENSG00000215612.8 | H6 family homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMX1 | hg38_v1_chr4_-_8871817_8871857 | 0.06 | 7.3e-01 | Click! |
Activity profile of HMX1 motif
Sorted Z-values of HMX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.9 | 2.8 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.9 | 6.3 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.8 | 2.3 | GO:0071810 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.7 | 3.6 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.7 | 2.8 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.6 | 3.9 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.6 | 3.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.6 | 4.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.6 | 6.5 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.5 | 6.2 | GO:0030578 | PML body organization(GO:0030578) positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.5 | 3.1 | GO:0070889 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.5 | 1.4 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.5 | 1.9 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.5 | 1.4 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.5 | 2.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.4 | 1.3 | GO:1902214 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 0.4 | 2.2 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.4 | 5.1 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.4 | 4.2 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.4 | 1.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.4 | 3.3 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.4 | 2.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.4 | 1.4 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.4 | 1.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.4 | 3.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 3.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 4.9 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.3 | 2.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.3 | 4.6 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.3 | 3.8 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.3 | 0.9 | GO:0070377 | negative regulation of ERK5 cascade(GO:0070377) |
| 0.3 | 1.2 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.3 | 0.3 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.3 | 3.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.2 | 2.7 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 | 0.7 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.2 | 1.5 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.2 | 1.5 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.2 | 3.8 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 2.9 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.2 | 1.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.2 | 1.0 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.2 | 2.8 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 | 0.5 | GO:0032824 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.2 | 0.9 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.2 | 0.7 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.2 | 3.4 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.2 | 0.8 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.2 | 0.2 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.2 | 2.8 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 0.7 | GO:0045337 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.4 | GO:1902997 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 1.0 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.6 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.1 | 2.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.9 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.1 | 1.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.8 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 10.9 | GO:0043303 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.1 | 1.5 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.1 | 1.3 | GO:1905049 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) negative regulation of trophoblast cell migration(GO:1901164) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.1 | 3.7 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 1.9 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 3.0 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 0.8 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 4.0 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.8 | GO:1905123 | regulation of glucosylceramidase activity(GO:1905123) |
| 0.1 | 1.0 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.7 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 1.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.3 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 1.1 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.1 | 3.0 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.4 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.4 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.1 | 0.3 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.1 | 1.9 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.1 | 0.5 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 1.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.6 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 1.2 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.1 | 1.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.9 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 2.0 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 3.4 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 | 7.0 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 2.2 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 1.6 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.8 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 1.0 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 3.8 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 1.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.7 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 1.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 3.0 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.1 | 0.4 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.6 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 1.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.2 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 2.0 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 2.2 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.5 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.3 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 2.0 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 10.1 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.7 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 1.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 7.7 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 1.3 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.2 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.0 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.9 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 1.7 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.4 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.2 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 1.6 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.3 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 1.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 1.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 2.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 1.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 1.3 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 1.7 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 1.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 3.0 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.9 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 2.7 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 1.0 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 1.0 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 1.1 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 1.0 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 1.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.2 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.5 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 1.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.6 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.2 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 2.3 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 1.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 3.4 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 1.3 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 2.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.4 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.5 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.6 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.5 | 1.9 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.5 | 3.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.4 | 3.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.4 | 8.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.4 | 2.3 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.4 | 2.9 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.3 | 2.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 11.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 0.8 | GO:1905103 | integral component of lysosomal membrane(GO:1905103) |
| 0.3 | 1.0 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.2 | 3.7 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.2 | 0.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 4.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 3.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 4.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 3.0 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.2 | 0.7 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 1.0 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.1 | 1.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 3.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 2.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 2.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.4 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.1 | 0.5 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 0.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 2.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 4.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 19.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 3.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 2.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 7.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.5 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.4 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 2.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 3.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 7.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 3.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.4 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 1.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 1.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 1.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 2.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 2.0 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 2.8 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 7.6 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.6 | 1.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.5 | 2.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.4 | 9.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 3.9 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 2.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.4 | 1.8 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.4 | 1.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.3 | 3.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.3 | 1.4 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.3 | 1.3 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.3 | 1.8 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.3 | 3.3 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.3 | 1.2 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.3 | 3.8 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 1.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 2.9 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.3 | 4.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.3 | 1.0 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.2 | 0.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 1.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 1.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 2.9 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 1.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 0.6 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.2 | 1.4 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 0.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 1.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 2.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 0.8 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.2 | 1.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.2 | 0.8 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 1.7 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 0.7 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 4.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 3.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.9 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.1 | 10.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.5 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 3.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 4.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 1.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.9 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 6.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 3.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.8 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.3 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.3 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 1.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.6 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.1 | 2.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.3 | GO:0004577 | N-acetylglucosaminyldiphosphodolichol N-acetylglucosaminyltransferase activity(GO:0004577) |
| 0.1 | 6.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.7 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 1.6 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.7 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 2.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 7.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 2.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 4.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.6 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 1.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 2.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 1.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 1.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 6.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 4.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 2.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 5.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 4.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.9 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.2 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 1.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 5.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.0 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 1.0 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 4.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 1.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 5.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 12.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.2 | 3.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 4.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 10.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 19.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 12.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 6.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 3.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 3.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 4.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 5.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 6.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 3.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 2.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.9 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 5.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 2.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 5.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.9 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.2 | 5.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 1.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.2 | 6.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 3.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 2.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 5.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.9 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 1.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 0.9 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 3.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 4.5 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 2.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.6 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 3.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 3.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 3.6 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 1.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 7.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 3.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.2 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.7 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 1.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.2 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 2.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.7 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.0 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.5 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |