Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for HOXA10_HOXB9
Z-value: 0.91
Transcription factors associated with HOXA10_HOXB9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
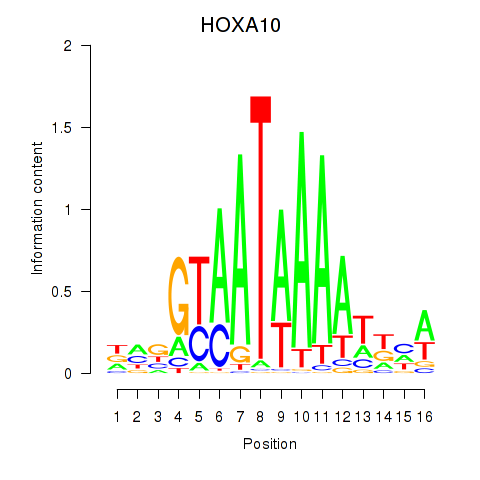
HOXA10
|
ENSG00000253293.5 | homeobox A10 |
|
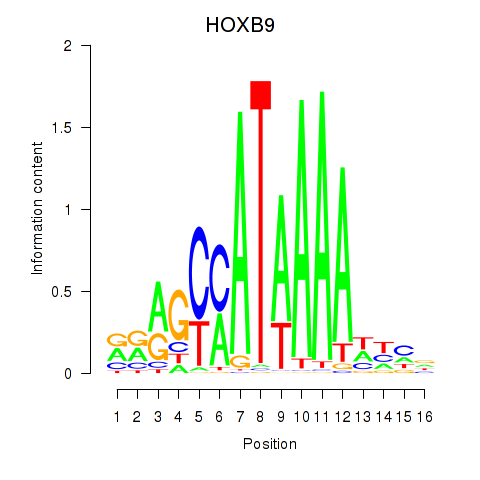
HOXB9
|
ENSG00000170689.10 | homeobox B9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB9 | hg38_v1_chr17_-_48626325_48626379 | 0.35 | 5.2e-02 | Click! |
| HOXA10 | hg38_v1_chr7_-_27180230_27180272 | -0.09 | 6.2e-01 | Click! |
Activity profile of HOXA10_HOXB9 motif
Sorted Z-values of HOXA10_HOXB9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_129144691 | 4.56 |
ENST00000486685.3
|
TSPAN33
|
tetraspanin 33 |
| chr4_-_71784046 | 4.45 |
ENST00000513476.5
ENST00000273951.13 |
GC
|
GC vitamin D binding protein |
| chr9_-_101435760 | 3.37 |
ENST00000647789.2
ENST00000616752.1 |
ALDOB
|
aldolase, fructose-bisphosphate B |
| chr4_+_154563003 | 3.05 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr9_-_101442403 | 2.43 |
ENST00000648758.1
|
ALDOB
|
aldolase, fructose-bisphosphate B |
| chr3_-_58210961 | 2.25 |
ENST00000486455.5
ENST00000394549.7 |
DNASE1L3
|
deoxyribonuclease 1 like 3 |
| chr10_-_72088533 | 1.96 |
ENST00000373109.7
|
SPOCK2
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 2 |
| chr15_+_48191648 | 1.93 |
ENST00000646012.1
ENST00000561127.5 ENST00000647546.1 ENST00000559641.5 ENST00000417307.3 |
SLC12A1
CTXN2
|
solute carrier family 12 member 1 cortexin 2 |
| chr10_-_72088504 | 1.90 |
ENST00000536168.2
|
SPOCK2
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 2 |
| chr10_+_94938649 | 1.83 |
ENST00000461906.1
ENST00000260682.8 |
CYP2C9
|
cytochrome P450 family 2 subfamily C member 9 |
| chr4_+_67558719 | 1.75 |
ENST00000265404.7
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr19_+_47994625 | 1.61 |
ENST00000339841.7
|
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr19_+_47994696 | 1.60 |
ENST00000596043.5
ENST00000597519.5 |
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr20_-_7940444 | 1.51 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1 |
| chr12_+_69825221 | 1.47 |
ENST00000552032.7
|
MYRFL
|
myelin regulatory factor like |
| chr2_+_233760265 | 1.44 |
ENST00000305208.10
ENST00000360418.4 |
UGT1A1
|
UDP glucuronosyltransferase family 1 member A1 |
| chr17_-_40665121 | 1.43 |
ENST00000394052.5
|
KRT222
|
keratin 222 |
| chr12_+_20810698 | 1.41 |
ENST00000540853.5
ENST00000381545.8 |
SLCO1B3
|
solute carrier organic anion transporter family member 1B3 |
| chr16_+_66579448 | 1.40 |
ENST00000379486.6
ENST00000268595.3 |
CMTM2
|
CKLF like MARVEL transmembrane domain containing 2 |
| chr16_+_72056153 | 1.38 |
ENST00000576168.6
ENST00000567185.7 ENST00000567612.2 |
HP
|
haptoglobin |
| chr9_-_101442372 | 1.36 |
ENST00000648423.1
|
ALDOB
|
aldolase, fructose-bisphosphate B |
| chr1_+_151762899 | 1.27 |
ENST00000635322.1
ENST00000321531.10 |
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr20_+_4721901 | 1.24 |
ENST00000305817.3
|
PRND
|
prion like protein doppel |
| chr13_+_108269629 | 1.22 |
ENST00000430559.5
ENST00000375887.9 |
TNFSF13B
|
TNF superfamily member 13b |
| chr10_+_133347347 | 1.18 |
ENST00000463201.2
ENST00000433452.6 |
PRAP1
|
proline rich acidic protein 1 |
| chr15_+_40594001 | 1.12 |
ENST00000346991.9
ENST00000528975.5 |
KNL1
|
kinetochore scaffold 1 |
| chr10_-_95069489 | 1.09 |
ENST00000371270.6
ENST00000535898.5 ENST00000623108.3 |
CYP2C8
|
cytochrome P450 family 2 subfamily C member 8 |
| chr12_-_89656093 | 1.09 |
ENST00000359142.7
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr20_+_44355692 | 1.05 |
ENST00000316673.8
ENST00000609795.5 ENST00000457232.5 ENST00000609262.5 |
HNF4A
|
hepatocyte nuclear factor 4 alpha |
| chr12_-_89656051 | 1.05 |
ENST00000261173.6
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr10_-_127892930 | 1.05 |
ENST00000368671.4
|
CLRN3
|
clarin 3 |
| chr1_+_244352627 | 1.00 |
ENST00000366537.5
ENST00000308105.5 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr15_+_40594241 | 1.00 |
ENST00000532056.5
ENST00000527044.5 ENST00000399668.7 |
KNL1
|
kinetochore scaffold 1 |
| chr10_+_92691813 | 0.99 |
ENST00000472590.6
|
HHEX
|
hematopoietically expressed homeobox |
| chr4_-_68951763 | 0.99 |
ENST00000251566.9
|
UGT2A3
|
UDP glucuronosyltransferase family 2 member A3 |
| chr3_-_123991352 | 0.96 |
ENST00000184183.8
|
ROPN1
|
rhophilin associated tail protein 1 |
| chr1_+_241532121 | 0.93 |
ENST00000366558.7
|
KMO
|
kynurenine 3-monooxygenase |
| chr14_+_22163226 | 0.92 |
ENST00000390458.3
|
TRAV29DV5
|
T cell receptor alpha variable 29/delta variable 5 |
| chr1_+_151766655 | 0.90 |
ENST00000400999.7
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr12_+_69825273 | 0.90 |
ENST00000547771.6
|
MYRFL
|
myelin regulatory factor like |
| chr2_-_157325808 | 0.89 |
ENST00000410096.6
ENST00000420719.6 ENST00000409216.5 ENST00000419116.2 |
ERMN
|
ermin |
| chr14_-_20590823 | 0.85 |
ENST00000556526.1
|
RNASE12
|
ribonuclease A family member 12 (inactive) |
| chr9_+_92974476 | 0.82 |
ENST00000337352.10
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr16_+_56191476 | 0.82 |
ENST00000262493.12
|
GNAO1
|
G protein subunit alpha o1 |
| chr1_-_120054225 | 0.79 |
ENST00000602566.6
|
NOTCH2
|
notch receptor 2 |
| chr4_+_186266183 | 0.79 |
ENST00000403665.7
ENST00000492972.6 ENST00000264692.8 |
F11
|
coagulation factor XI |
| chr13_+_108269880 | 0.75 |
ENST00000542136.1
|
TNFSF13B
|
TNF superfamily member 13b |
| chr8_-_80171496 | 0.75 |
ENST00000379096.9
ENST00000518937.6 |
TPD52
|
tumor protein D52 |
| chr19_-_19515542 | 0.73 |
ENST00000585580.4
|
TSSK6
|
testis specific serine kinase 6 |
| chrX_-_81201886 | 0.73 |
ENST00000451455.1
ENST00000358130.7 ENST00000436386.5 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr7_-_143408848 | 0.73 |
ENST00000275815.4
|
EPHA1
|
EPH receptor A1 |
| chr12_+_25052512 | 0.73 |
ENST00000557489.5
ENST00000354454.7 ENST00000536173.5 |
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr1_+_241532370 | 0.71 |
ENST00000366559.9
ENST00000366557.8 |
KMO
|
kynurenine 3-monooxygenase |
| chr12_+_20815672 | 0.71 |
ENST00000261196.6
ENST00000381541.7 ENST00000540229.1 |
SLCO1B3
SLCO1B3-SLCO1B7
|
solute carrier organic anion transporter family member 1B3 SLCO1B3-SLCO1B7 readthrough |
| chr1_+_84164891 | 0.70 |
ENST00000413538.5
ENST00000417530.5 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr2_+_196713117 | 0.70 |
ENST00000409270.5
|
CCDC150
|
coiled-coil domain containing 150 |
| chr10_-_14532678 | 0.69 |
ENST00000489100.5
|
FAM107B
|
family with sequence similarity 107 member B |
| chr9_-_101383558 | 0.69 |
ENST00000674556.1
|
BAAT
|
bile acid-CoA:amino acid N-acyltransferase |
| chr16_-_1884231 | 0.67 |
ENST00000563416.3
ENST00000633813.1 ENST00000470044.5 |
LINC00254
MEIOB
|
long intergenic non-protein coding RNA 254 meiosis specific with OB-fold |
| chr3_+_125969152 | 0.67 |
ENST00000251776.8
ENST00000504401.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr9_+_101185029 | 0.67 |
ENST00000395056.2
|
PLPPR1
|
phospholipid phosphatase related 1 |
| chr1_+_84164962 | 0.66 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr11_-_26723361 | 0.65 |
ENST00000533617.5
|
SLC5A12
|
solute carrier family 5 member 12 |
| chr21_-_14210884 | 0.64 |
ENST00000679868.1
ENST00000400211.3 ENST00000680801.1 ENST00000536861.6 ENST00000614229.5 |
LIPI
|
lipase I |
| chr1_+_173635332 | 0.63 |
ENST00000417563.3
|
TEX50
|
testis expressed 50 |
| chr1_+_154429315 | 0.62 |
ENST00000476006.5
|
IL6R
|
interleukin 6 receptor |
| chr15_-_55249029 | 0.61 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr10_-_97270638 | 0.60 |
ENST00000371027.5
|
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr14_-_60486021 | 0.60 |
ENST00000555476.5
ENST00000321731.8 |
C14orf39
|
chromosome 14 open reading frame 39 |
| chrX_+_37780049 | 0.59 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain |
| chr3_+_125969172 | 0.59 |
ENST00000514116.6
ENST00000513830.5 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr6_+_6588082 | 0.58 |
ENST00000379953.6
|
LY86
|
lymphocyte antigen 86 |
| chr2_-_51032151 | 0.57 |
ENST00000628515.2
|
NRXN1
|
neurexin 1 |
| chr12_-_81598360 | 0.57 |
ENST00000333447.11
ENST00000407050.8 |
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chrX_+_106693838 | 0.57 |
ENST00000324342.7
|
RNF128
|
ring finger protein 128 |
| chrX_+_41689006 | 0.56 |
ENST00000378138.5
ENST00000620846.1 ENST00000649219.1 |
GPR34
|
G protein-coupled receptor 34 |
| chr14_+_21965451 | 0.56 |
ENST00000390442.3
|
TRAV12-3
|
T cell receptor alpha variable 12-3 |
| chr1_-_186461089 | 0.55 |
ENST00000391997.3
|
PDC
|
phosducin |
| chr9_-_111759508 | 0.55 |
ENST00000394777.8
ENST00000394779.7 |
SHOC1
|
shortage in chiasmata 1 |
| chr1_+_51102221 | 0.55 |
ENST00000648827.1
ENST00000371759.7 |
C1orf185
|
chromosome 1 open reading frame 185 |
| chr9_+_102995308 | 0.55 |
ENST00000612124.4
ENST00000374798.8 ENST00000487798.5 |
CYLC2
|
cylicin 2 |
| chr8_-_70245343 | 0.55 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr10_+_92691897 | 0.54 |
ENST00000492654.3
|
HHEX
|
hematopoietically expressed homeobox |
| chr12_+_25052634 | 0.54 |
ENST00000548766.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr4_+_87975667 | 0.52 |
ENST00000237623.11
ENST00000682655.1 ENST00000508233.6 ENST00000360804.4 ENST00000395080.8 |
SPP1
|
secreted phosphoprotein 1 |
| chr1_+_192158448 | 0.52 |
ENST00000367460.4
|
RGS18
|
regulator of G protein signaling 18 |
| chr8_+_75539893 | 0.52 |
ENST00000674002.1
|
HNF4G
|
hepatocyte nuclear factor 4 gamma |
| chr2_+_37950432 | 0.52 |
ENST00000407257.5
ENST00000417700.6 ENST00000234195.7 ENST00000442857.5 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr4_+_71062642 | 0.51 |
ENST00000649996.1
|
SLC4A4
|
solute carrier family 4 member 4 |
| chr21_+_33025927 | 0.49 |
ENST00000430860.1
ENST00000382357.4 ENST00000333337.3 |
OLIG2
|
oligodendrocyte transcription factor 2 |
| chr20_+_10035029 | 0.49 |
ENST00000378380.4
|
ANKEF1
|
ankyrin repeat and EF-hand domain containing 1 |
| chr11_-_5516690 | 0.49 |
ENST00000380184.2
|
UBQLNL
|
ubiquilin like |
| chr20_-_31406207 | 0.48 |
ENST00000376314.3
|
DEFB121
|
defensin beta 121 |
| chr9_+_131190119 | 0.48 |
ENST00000483497.6
|
NUP214
|
nucleoporin 214 |
| chr12_-_81598332 | 0.48 |
ENST00000443686.7
|
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr4_+_94207845 | 0.48 |
ENST00000457823.6
ENST00000354268.9 |
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_+_37453914 | 0.48 |
ENST00000381980.9
ENST00000508175.5 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr12_+_78863962 | 0.47 |
ENST00000393240.7
|
SYT1
|
synaptotagmin 1 |
| chr8_+_75539862 | 0.46 |
ENST00000396423.4
|
HNF4G
|
hepatocyte nuclear factor 4 gamma |
| chr16_-_21652598 | 0.46 |
ENST00000569602.1
ENST00000268389.6 |
IGSF6
|
immunoglobulin superfamily member 6 |
| chr14_-_67533720 | 0.46 |
ENST00000554278.6
|
TMEM229B
|
transmembrane protein 229B |
| chr20_+_10034963 | 0.45 |
ENST00000378392.6
|
ANKEF1
|
ankyrin repeat and EF-hand domain containing 1 |
| chr5_-_36001006 | 0.45 |
ENST00000625798.2
|
UGT3A1
|
UDP glycosyltransferase family 3 member A1 |
| chr17_-_10657302 | 0.44 |
ENST00000583535.6
|
MYH3
|
myosin heavy chain 3 |
| chr2_-_157325659 | 0.43 |
ENST00000409925.1
|
ERMN
|
ermin |
| chr9_-_71121596 | 0.43 |
ENST00000377110.9
ENST00000377111.8 ENST00000677713.2 |
TRPM3
|
transient receptor potential cation channel subfamily M member 3 |
| chr21_+_34364003 | 0.43 |
ENST00000290310.4
|
KCNE2
|
potassium voltage-gated channel subfamily E regulatory subunit 2 |
| chrY_+_18546691 | 0.42 |
ENST00000309834.8
ENST00000307393.3 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor Y-linked 1 |
| chr1_+_51102238 | 0.42 |
ENST00000467127.5
|
C1orf185
|
chromosome 1 open reading frame 185 |
| chr2_+_31234144 | 0.42 |
ENST00000322054.10
|
EHD3
|
EH domain containing 3 |
| chr5_-_131165231 | 0.42 |
ENST00000675100.1
ENST00000304043.10 ENST00000513012.2 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr10_-_125816510 | 0.42 |
ENST00000650587.1
|
UROS
|
uroporphyrinogen III synthase |
| chr2_+_113127588 | 0.41 |
ENST00000409930.4
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr7_-_123199960 | 0.41 |
ENST00000194130.7
|
SLC13A1
|
solute carrier family 13 member 1 |
| chr5_+_140882116 | 0.41 |
ENST00000289272.3
ENST00000409494.5 ENST00000617769.1 |
PCDHA13
|
protocadherin alpha 13 |
| chr17_-_31297231 | 0.41 |
ENST00000247271.5
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr10_-_113854368 | 0.41 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr4_-_68670648 | 0.41 |
ENST00000338206.6
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15 |
| chr12_+_40310431 | 0.40 |
ENST00000681696.1
|
LRRK2
|
leucine rich repeat kinase 2 |
| chr12_-_49187369 | 0.40 |
ENST00000547939.6
|
TUBA1A
|
tubulin alpha 1a |
| chr16_-_48247533 | 0.40 |
ENST00000356608.7
ENST00000569991.1 |
ABCC11
|
ATP binding cassette subfamily C member 11 |
| chr13_-_102759059 | 0.39 |
ENST00000322527.4
|
CCDC168
|
coiled-coil domain containing 168 |
| chr3_+_101827982 | 0.39 |
ENST00000461724.5
ENST00000483180.5 ENST00000394054.6 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr16_-_30610342 | 0.39 |
ENST00000287461.8
|
ZNF689
|
zinc finger protein 689 |
| chr6_-_69699124 | 0.38 |
ENST00000651675.1
|
LMBRD1
|
LMBR1 domain containing 1 |
| chr2_-_51032091 | 0.38 |
ENST00000637511.1
ENST00000405472.7 ENST00000405581.3 ENST00000401669.7 |
NRXN1
|
neurexin 1 |
| chr5_-_131165272 | 0.38 |
ENST00000675491.1
ENST00000506908.2 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr17_-_10549612 | 0.38 |
ENST00000532183.6
ENST00000397183.6 ENST00000420805.1 |
MYH2
|
myosin heavy chain 2 |
| chr7_-_78771265 | 0.37 |
ENST00000630991.2
ENST00000629359.2 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr7_-_50565381 | 0.37 |
ENST00000444124.7
|
DDC
|
dopa decarboxylase |
| chr22_-_23580223 | 0.37 |
ENST00000249053.3
ENST00000330377.3 ENST00000438703.1 |
IGLL1
|
immunoglobulin lambda like polypeptide 1 |
| chr6_+_87344812 | 0.37 |
ENST00000388923.5
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr4_+_70397931 | 0.37 |
ENST00000399575.7
|
OPRPN
|
opiorphin prepropeptide |
| chr6_-_8102481 | 0.36 |
ENST00000502429.5
ENST00000429723.6 ENST00000379715.10 ENST00000507463.1 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr2_+_73784189 | 0.36 |
ENST00000409561.1
|
C2orf78
|
chromosome 2 open reading frame 78 |
| chr5_-_161546970 | 0.36 |
ENST00000675303.1
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr1_-_161238085 | 0.36 |
ENST00000512372.5
ENST00000437437.6 ENST00000412844.6 ENST00000428574.6 ENST00000442691.6 ENST00000505005.5 ENST00000508740.5 ENST00000367981.7 ENST00000502985.5 ENST00000504010.5 ENST00000508387.5 ENST00000511676.5 ENST00000511748.5 ENST00000511944.5 ENST00000515621.5 ENST00000367984.8 ENST00000367985.7 |
NR1I3
|
nuclear receptor subfamily 1 group I member 3 |
| chr13_-_46438190 | 0.35 |
ENST00000409879.6
|
RUBCNL
|
rubicon like autophagy enhancer |
| chr13_+_24270681 | 0.35 |
ENST00000343003.10
ENST00000399949.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr17_-_10549652 | 0.35 |
ENST00000245503.10
|
MYH2
|
myosin heavy chain 2 |
| chr16_+_76277393 | 0.35 |
ENST00000611870.5
|
CNTNAP4
|
contactin associated protein family member 4 |
| chr9_+_60914407 | 0.34 |
ENST00000437823.5
|
SPATA31A5
|
SPATA31 subfamily A member 5 |
| chr15_-_83283449 | 0.34 |
ENST00000569704.2
|
BNC1
|
basonuclin 1 |
| chr12_+_25052220 | 0.34 |
ENST00000550945.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chrX_-_54043927 | 0.34 |
ENST00000415025.5
ENST00000338946.10 |
PHF8
|
PHD finger protein 8 |
| chrY_-_18773686 | 0.34 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor Y-linked 2 |
| chr14_+_22168387 | 0.33 |
ENST00000557168.1
|
TRAV30
|
T cell receptor alpha variable 30 |
| chrX_-_135781729 | 0.33 |
ENST00000617203.1
|
CT45A5
|
cancer/testis antigen family 45 member A5 |
| chr2_-_240820945 | 0.33 |
ENST00000428768.2
ENST00000650053.1 ENST00000650130.1 |
KIF1A
|
kinesin family member 1A |
| chr14_+_22086401 | 0.33 |
ENST00000390451.2
|
TRAV23DV6
|
T cell receptor alpha variable 23/delta variable 6 |
| chr17_-_75667088 | 0.33 |
ENST00000578201.5
ENST00000423245.6 ENST00000317905.10 |
RECQL5
|
RecQ like helicase 5 |
| chr5_-_20575850 | 0.32 |
ENST00000507958.5
|
CDH18
|
cadherin 18 |
| chr6_+_80106623 | 0.32 |
ENST00000369760.8
ENST00000356489.9 |
BCKDHB
|
branched chain keto acid dehydrogenase E1 subunit beta |
| chr3_-_15798184 | 0.32 |
ENST00000624145.3
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr10_+_89392546 | 0.32 |
ENST00000546318.2
ENST00000371804.4 |
IFIT1
|
interferon induced protein with tetratricopeptide repeats 1 |
| chr1_+_44405164 | 0.32 |
ENST00000355387.6
|
RNF220
|
ring finger protein 220 |
| chr1_+_92168915 | 0.32 |
ENST00000637221.2
|
BTBD8
|
BTB domain containing 8 |
| chr19_+_20923222 | 0.32 |
ENST00000597314.5
ENST00000328178.13 ENST00000601924.5 |
ZNF85
|
zinc finger protein 85 |
| chr6_+_25754699 | 0.31 |
ENST00000439485.6
ENST00000377905.9 |
SLC17A4
|
solute carrier family 17 member 4 |
| chr2_-_175181663 | 0.31 |
ENST00000392541.3
ENST00000284727.9 ENST00000409194.5 |
ATP5MC3
|
ATP synthase membrane subunit c locus 3 |
| chr17_+_29941605 | 0.31 |
ENST00000394835.7
|
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr7_+_130344810 | 0.31 |
ENST00000497503.5
ENST00000463587.5 ENST00000461828.5 ENST00000474905.6 ENST00000494311.1 ENST00000466363.6 |
CPA5
|
carboxypeptidase A5 |
| chr9_-_130939205 | 0.30 |
ENST00000372338.9
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr6_-_109483208 | 0.30 |
ENST00000230122.4
|
ZBTB24
|
zinc finger and BTB domain containing 24 |
| chrX_-_54043143 | 0.30 |
ENST00000445025.1
ENST00000322659.12 |
PHF8
|
PHD finger protein 8 |
| chr12_-_81758665 | 0.30 |
ENST00000549325.5
ENST00000550584.6 |
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr16_-_9943182 | 0.30 |
ENST00000535259.6
|
GRIN2A
|
glutamate ionotropic receptor NMDA type subunit 2A |
| chr12_-_112006024 | 0.30 |
ENST00000550800.6
ENST00000550037.5 ENST00000549425.5 |
TMEM116
|
transmembrane protein 116 |
| chr1_-_206970457 | 0.30 |
ENST00000324852.9
ENST00000450945.3 ENST00000400962.8 |
FCAMR
|
Fc fragment of IgA and IgM receptor |
| chr12_-_81758641 | 0.29 |
ENST00000552948.5
ENST00000548586.5 |
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr17_-_75667165 | 0.29 |
ENST00000584999.1
ENST00000420326.6 ENST00000340830.9 |
RECQL5
|
RecQ like helicase 5 |
| chr12_-_7503841 | 0.29 |
ENST00000359156.8
|
CD163
|
CD163 molecule |
| chr3_+_88338451 | 0.29 |
ENST00000637986.2
|
CSNKA2IP
|
casein kinase 2 subunit alpha' interacting protein |
| chr9_-_104536856 | 0.29 |
ENST00000641090.1
|
OR13C3
|
olfactory receptor family 13 subfamily C member 3 |
| chr3_+_125969214 | 0.29 |
ENST00000508088.1
|
ROPN1B
|
rhophilin associated tail protein 1B |
| chr14_+_30577752 | 0.29 |
ENST00000547532.5
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr15_-_50546660 | 0.29 |
ENST00000532404.6
ENST00000616326.1 |
USP50
|
ubiquitin specific peptidase 50 |
| chr3_+_138621207 | 0.29 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr10_-_16521871 | 0.29 |
ENST00000298943.4
|
C1QL3
|
complement C1q like 3 |
| chrX_+_48761743 | 0.29 |
ENST00000303227.11
|
GLOD5
|
glyoxalase domain containing 5 |
| chr6_+_158312459 | 0.28 |
ENST00000367097.8
|
TULP4
|
TUB like protein 4 |
| chr9_+_121121760 | 0.28 |
ENST00000373847.5
|
CNTRL
|
centriolin |
| chr18_+_58196736 | 0.28 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr19_+_3185911 | 0.28 |
ENST00000246117.9
ENST00000588428.5 |
NCLN
|
nicalin |
| chr19_+_4402615 | 0.28 |
ENST00000301280.10
|
CHAF1A
|
chromatin assembly factor 1 subunit A |
| chr13_-_46052712 | 0.28 |
ENST00000242848.8
ENST00000679008.1 ENST00000282007.7 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr14_+_22207502 | 0.28 |
ENST00000390461.2
|
TRAV34
|
T cell receptor alpha variable 34 |
| chr15_+_63122561 | 0.28 |
ENST00000557972.1
|
LACTB
|
lactamase beta |
| chr20_+_38066060 | 0.27 |
ENST00000614670.1
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr11_+_63838902 | 0.27 |
ENST00000377810.8
|
MARK2
|
microtubule affinity regulating kinase 2 |
| chr12_+_25052732 | 0.27 |
ENST00000547044.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr7_-_6026552 | 0.27 |
ENST00000422786.1
|
EIF2AK1
|
eukaryotic translation initiation factor 2 alpha kinase 1 |
| chr3_+_172754457 | 0.26 |
ENST00000441497.6
|
ECT2
|
epithelial cell transforming 2 |
| chr8_-_21812320 | 0.26 |
ENST00000517328.5
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr12_+_75334655 | 0.26 |
ENST00000378695.9
|
GLIPR1L1
|
GLIPR1 like 1 |
| chr17_+_59729702 | 0.26 |
ENST00000587259.5
|
VMP1
|
vacuole membrane protein 1 |
| chr7_-_57132425 | 0.26 |
ENST00000319636.10
|
ZNF479
|
zinc finger protein 479 |
| chr19_+_4402685 | 0.26 |
ENST00000585854.1
|
CHAF1A
|
chromatin assembly factor 1 subunit A |
| chr5_-_161546708 | 0.26 |
ENST00000393959.6
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr11_-_3379212 | 0.26 |
ENST00000429541.6
ENST00000532539.1 ENST00000343338.11 ENST00000620374.4 |
ZNF195
|
zinc finger protein 195 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA10_HOXB9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.7 | 7.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.6 | 1.8 | GO:1903970 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.5 | 1.5 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.5 | 2.0 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.5 | 1.4 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.4 | 1.4 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.3 | 1.6 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.3 | 2.9 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.2 | 2.1 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.2 | 3.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 0.8 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.2 | 0.6 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.2 | 1.5 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.2 | 3.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 0.5 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.7 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 0.5 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 1.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 4.5 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.4 | GO:1903123 | regulation of thioredoxin peroxidase activity(GO:1903123) negative regulation of thioredoxin peroxidase activity(GO:1903124) negative regulation of thioredoxin peroxidase activity by peptidyl-threonine phosphorylation(GO:1903125) Wnt signalosome assembly(GO:1904887) negative regulation of peroxidase activity(GO:2000469) |
| 0.1 | 0.6 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 0.4 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.1 | 0.6 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 2.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.6 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.5 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 0.4 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 2.3 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 3.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.6 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.4 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.1 | 0.3 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.3 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.5 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 1.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.1 | 0.3 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.1 | 0.4 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.1 | 1.0 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 0.3 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 1.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.3 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 0.4 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.6 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 1.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 0.8 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.7 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 1.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 1.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.7 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.0 | 0.8 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.4 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 2.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.7 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.6 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 1.0 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.8 | GO:0060413 | atrial septum morphogenesis(GO:0060413) Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.4 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.4 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.4 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.4 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.3 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.1 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.2 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.0 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.5 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.2 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.3 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.2 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.4 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.9 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.6 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.7 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.8 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.0 | 3.9 | GO:0090002 | establishment of protein localization to plasma membrane(GO:0090002) |
| 0.0 | 0.2 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 3.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 3.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 0.5 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.1 | 7.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.4 | GO:0099400 | cytoplasmic side of mitochondrial outer membrane(GO:0032473) caveola neck(GO:0099400) |
| 0.1 | 1.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 1.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.5 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 2.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.5 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 1.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.4 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.4 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 0.5 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 1.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 2.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 1.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 5.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.0 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 2.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 1.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.2 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 1.1 | 4.5 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.7 | 2.2 | GO:0015489 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.6 | 1.9 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.6 | 2.9 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.5 | 1.5 | GO:0052853 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.2 | 0.7 | GO:0052815 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.2 | 1.8 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 1.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 0.5 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.2 | 0.6 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 0.7 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 3.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.4 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.4 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) peroxidase inhibitor activity(GO:0036479) |
| 0.1 | 0.8 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 1.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.3 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.4 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.5 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 2.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.6 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.7 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.7 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 0.8 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.1 | 3.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.2 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.1 | 0.2 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.1 | 2.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.4 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 1.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.4 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.4 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 2.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.7 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 1.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.9 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.4 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.8 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.4 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 1.5 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 2.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 2.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 3.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 8.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 4.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.8 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 2.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |