Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
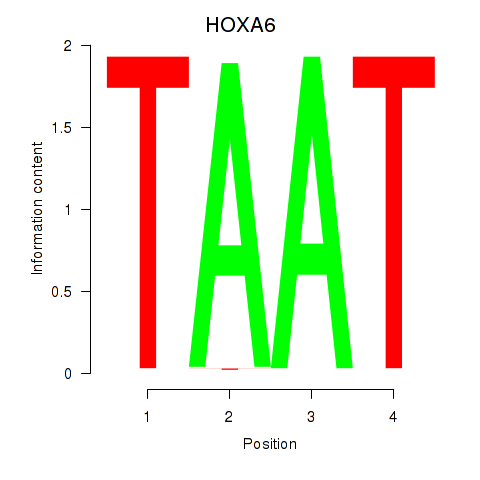
Results for HOXA6
Z-value: 1.24
Transcription factors associated with HOXA6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA6
|
ENSG00000106006.6 | homeobox A6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA6 | hg38_v1_chr7_-_27147774_27147774 | -0.00 | 9.9e-01 | Click! |
Activity profile of HOXA6 motif
Sorted Z-values of HOXA6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_73404255 | 5.83 |
ENST00000621628.4
ENST00000621085.4 ENST00000415165.6 ENST00000295897.9 ENST00000503124.5 ENST00000509063.5 ENST00000401494.7 |
ALB
|
albumin |
| chr10_+_113125536 | 4.86 |
ENST00000349937.7
|
TCF7L2
|
transcription factor 7 like 2 |
| chr4_-_71784046 | 4.74 |
ENST00000513476.5
ENST00000273951.13 |
GC
|
GC vitamin D binding protein |
| chr15_+_58138368 | 4.55 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chr3_+_41194741 | 4.32 |
ENST00000643541.1
ENST00000426215.5 ENST00000645210.1 ENST00000646381.1 ENST00000405570.6 ENST00000642248.1 ENST00000433400.6 |
CTNNB1
|
catenin beta 1 |
| chr14_-_94770102 | 4.15 |
ENST00000238558.5
|
GSC
|
goosecoid homeobox |
| chr17_-_40937445 | 3.99 |
ENST00000436344.7
ENST00000485751.1 |
KRT23
|
keratin 23 |
| chr1_+_186296267 | 3.95 |
ENST00000533951.5
ENST00000367482.8 ENST00000635041.1 ENST00000367483.8 ENST00000367485.4 ENST00000445192.7 |
PRG4
|
proteoglycan 4 |
| chr1_+_167329044 | 3.94 |
ENST00000367862.9
|
POU2F1
|
POU class 2 homeobox 1 |
| chr15_-_70097874 | 3.80 |
ENST00000557997.5
ENST00000317509.12 ENST00000451782.7 ENST00000627388.2 |
TLE3
|
TLE family member 3, transcriptional corepressor |
| chr15_+_58138169 | 3.63 |
ENST00000558772.5
|
AQP9
|
aquaporin 9 |
| chr3_-_71304741 | 3.52 |
ENST00000484350.5
|
FOXP1
|
forkhead box P1 |
| chr1_+_167329148 | 3.42 |
ENST00000420254.7
|
POU2F1
|
POU class 2 homeobox 1 |
| chr6_+_21593742 | 3.20 |
ENST00000244745.4
|
SOX4
|
SRY-box transcription factor 4 |
| chr16_-_51151238 | 3.07 |
ENST00000566102.1
|
SALL1
|
spalt like transcription factor 1 |
| chr17_-_48615261 | 3.00 |
ENST00000239144.5
|
HOXB8
|
homeobox B8 |
| chr17_-_40937641 | 2.88 |
ENST00000209718.8
|
KRT23
|
keratin 23 |
| chr2_-_176083913 | 2.84 |
ENST00000308618.4
|
EVX2
|
even-skipped homeobox 2 |
| chr16_-_51151259 | 2.77 |
ENST00000251020.9
|
SALL1
|
spalt like transcription factor 1 |
| chr17_-_48185179 | 2.75 |
ENST00000579336.1
|
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr20_+_6007245 | 2.70 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr11_+_31650024 | 2.69 |
ENST00000638317.1
|
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr7_-_27165457 | 2.66 |
ENST00000673744.1
ENST00000673917.1 |
HOXA9
|
homeobox A9 |
| chr17_-_48593748 | 2.64 |
ENST00000239151.6
|
HOXB5
|
homeobox B5 |
| chr9_-_121050264 | 2.63 |
ENST00000223642.3
|
C5
|
complement C5 |
| chr6_+_104957099 | 2.58 |
ENST00000345080.5
|
LIN28B
|
lin-28 homolog B |
| chr5_-_77638713 | 2.58 |
ENST00000306422.5
|
OTP
|
orthopedia homeobox |
| chr2_+_170715317 | 2.56 |
ENST00000375281.4
|
SP5
|
Sp5 transcription factor |
| chr10_+_116324440 | 2.56 |
ENST00000333254.4
|
CCDC172
|
coiled-coil domain containing 172 |
| chr18_-_32772879 | 2.56 |
ENST00000358095.4
ENST00000359358.9 |
KLHL14
|
kelch like family member 14 |
| chr10_-_101229449 | 2.49 |
ENST00000370193.4
|
LBX1
|
ladybird homeobox 1 |
| chr7_-_27165517 | 2.48 |
ENST00000396345.1
ENST00000343483.7 |
HOXA9
|
homeobox A9 |
| chr3_-_158106408 | 2.43 |
ENST00000483851.7
|
SHOX2
|
short stature homeobox 2 |
| chr2_-_144431001 | 2.30 |
ENST00000636413.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_-_23951020 | 2.27 |
ENST00000441133.2
ENST00000545921.5 |
SOX5
|
SRY-box transcription factor 5 |
| chr12_-_102950835 | 2.24 |
ENST00000546844.1
|
PAH
|
phenylalanine hydroxylase |
| chr4_-_115113614 | 2.19 |
ENST00000264363.7
|
NDST4
|
N-deacetylase and N-sulfotransferase 4 |
| chr6_-_32190170 | 2.16 |
ENST00000375050.6
|
PBX2
|
PBX homeobox 2 |
| chr6_+_130018565 | 2.13 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr7_-_78489900 | 2.12 |
ENST00000636039.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr17_-_40799939 | 2.12 |
ENST00000306658.8
|
KRT28
|
keratin 28 |
| chr11_-_8268716 | 2.09 |
ENST00000428101.6
|
LMO1
|
LIM domain only 1 |
| chr15_-_70097852 | 2.04 |
ENST00000559191.5
|
TLE3
|
TLE family member 3, transcriptional corepressor |
| chr14_-_34875348 | 2.04 |
ENST00000360310.6
|
BAZ1A
|
bromodomain adjacent to zinc finger domain 1A |
| chr3_-_57199938 | 2.03 |
ENST00000473921.2
ENST00000295934.8 |
HESX1
|
HESX homeobox 1 |
| chr4_-_115113822 | 2.02 |
ENST00000613194.4
|
NDST4
|
N-deacetylase and N-sulfotransferase 4 |
| chr7_-_5423543 | 2.01 |
ENST00000399537.8
|
TNRC18
|
trinucleotide repeat containing 18 |
| chr12_-_89352487 | 1.98 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr1_+_167220870 | 1.97 |
ENST00000367866.7
ENST00000429375.6 ENST00000541643.7 |
POU2F1
|
POU class 2 homeobox 1 |
| chr12_-_89352395 | 1.96 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr11_-_82845734 | 1.96 |
ENST00000681883.1
ENST00000680040.1 ENST00000681432.1 |
PRCP
|
prolylcarboxypeptidase |
| chr6_-_107824294 | 1.93 |
ENST00000369020.8
ENST00000369022.6 |
SCML4
|
Scm polycomb group protein like 4 |
| chr6_+_45328203 | 1.91 |
ENST00000371432.7
ENST00000647337.2 ENST00000371438.5 |
RUNX2
|
RUNX family transcription factor 2 |
| chr3_-_27722699 | 1.90 |
ENST00000461503.2
|
EOMES
|
eomesodermin |
| chr4_+_99574812 | 1.90 |
ENST00000422897.6
ENST00000265517.10 |
MTTP
|
microsomal triglyceride transfer protein |
| chr6_-_65707214 | 1.89 |
ENST00000370621.7
ENST00000393380.6 ENST00000503581.6 |
EYS
|
eyes shut homolog |
| chr14_-_68794597 | 1.82 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein like 1 |
| chr4_+_128809684 | 1.79 |
ENST00000226319.11
ENST00000511647.5 |
JADE1
|
jade family PHD finger 1 |
| chr9_+_2159850 | 1.78 |
ENST00000416751.2
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_-_78489955 | 1.78 |
ENST00000636178.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr17_-_48545077 | 1.76 |
ENST00000330070.6
|
HOXB2
|
homeobox B2 |
| chr11_+_128693887 | 1.75 |
ENST00000281428.12
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_+_99606833 | 1.73 |
ENST00000369215.5
|
PRDM13
|
PR/SET domain 13 |
| chr14_-_56816693 | 1.71 |
ENST00000673035.1
|
OTX2
|
orthodenticle homeobox 2 |
| chr13_-_74133892 | 1.70 |
ENST00000377669.7
|
KLF12
|
Kruppel like factor 12 |
| chr7_-_42152444 | 1.69 |
ENST00000479210.1
|
GLI3
|
GLI family zinc finger 3 |
| chr17_+_44957907 | 1.68 |
ENST00000678938.1
|
NMT1
|
N-myristoyltransferase 1 |
| chr11_+_128694052 | 1.68 |
ENST00000527786.7
ENST00000534087.3 |
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr16_+_8712943 | 1.61 |
ENST00000561870.5
ENST00000396600.6 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr9_+_2015275 | 1.60 |
ENST00000637103.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr12_-_23950906 | 1.59 |
ENST00000541847.5
|
SOX5
|
SRY-box transcription factor 5 |
| chr7_-_27102669 | 1.55 |
ENST00000222718.7
|
HOXA2
|
homeobox A2 |
| chrX_+_9463272 | 1.51 |
ENST00000407597.7
ENST00000380961.5 ENST00000424279.6 |
TBL1X
|
transducin beta like 1 X-linked |
| chr10_+_47322450 | 1.51 |
ENST00000581492.3
|
GDF2
|
growth differentiation factor 2 |
| chr12_-_102197827 | 1.49 |
ENST00000329406.5
|
PMCH
|
pro-melanin concentrating hormone |
| chr18_-_76495191 | 1.45 |
ENST00000443185.7
|
ZNF516
|
zinc finger protein 516 |
| chr18_+_31376777 | 1.43 |
ENST00000308128.9
ENST00000359747.4 |
DSG4
|
desmoglein 4 |
| chr6_+_144659814 | 1.43 |
ENST00000367525.3
|
UTRN
|
utrophin |
| chr2_-_181680490 | 1.40 |
ENST00000684145.1
ENST00000295108.4 ENST00000684079.1 ENST00000683430.1 |
CERKL
NEUROD1
|
ceramide kinase like neuronal differentiation 1 |
| chr20_-_32536383 | 1.40 |
ENST00000375678.7
|
NOL4L
|
nucleolar protein 4 like |
| chr6_-_24719146 | 1.40 |
ENST00000378119.9
|
C6orf62
|
chromosome 6 open reading frame 62 |
| chr14_+_71586261 | 1.39 |
ENST00000358550.6
|
SIPA1L1
|
signal induced proliferation associated 1 like 1 |
| chr2_+_176157293 | 1.37 |
ENST00000683222.1
|
HOXD3
|
homeobox D3 |
| chr7_+_18495697 | 1.33 |
ENST00000413380.5
ENST00000430454.5 |
HDAC9
|
histone deacetylase 9 |
| chr8_-_92095410 | 1.32 |
ENST00000518823.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr2_-_144521042 | 1.30 |
ENST00000637267.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr7_+_114414809 | 1.29 |
ENST00000350908.9
|
FOXP2
|
forkhead box P2 |
| chr1_-_205455954 | 1.29 |
ENST00000495594.2
ENST00000629624.2 |
LEMD1
BLACAT1
|
LEM domain containing 1 bladder cancer associated transcript 1 |
| chr4_+_159267737 | 1.29 |
ENST00000264431.8
|
RAPGEF2
|
Rap guanine nucleotide exchange factor 2 |
| chr5_-_91383310 | 1.27 |
ENST00000265138.4
|
ARRDC3
|
arrestin domain containing 3 |
| chr7_-_116030750 | 1.27 |
ENST00000265440.12
ENST00000320239.11 |
TFEC
|
transcription factor EC |
| chr4_-_112516176 | 1.25 |
ENST00000313341.4
|
NEUROG2
|
neurogenin 2 |
| chr15_+_80072559 | 1.25 |
ENST00000560228.5
ENST00000559835.5 ENST00000559775.5 ENST00000558688.5 ENST00000560392.5 ENST00000560976.5 ENST00000558272.5 ENST00000558390.5 |
ZFAND6
|
zinc finger AN1-type containing 6 |
| chr10_-_21517825 | 1.24 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr7_-_78489983 | 1.24 |
ENST00000519748.5
ENST00000634996.1 ENST00000637282.1 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr2_-_207165923 | 1.23 |
ENST00000309446.11
|
KLF7
|
Kruppel like factor 7 |
| chr7_-_41703062 | 1.23 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr14_+_22501542 | 1.22 |
ENST00000390498.1
|
TRAJ39
|
T cell receptor alpha joining 39 |
| chr9_+_2159672 | 1.22 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_+_76683779 | 1.22 |
ENST00000523885.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr20_-_51802509 | 1.22 |
ENST00000371539.7
ENST00000217086.9 |
SALL4
|
spalt like transcription factor 4 |
| chr7_-_5423826 | 1.22 |
ENST00000430969.6
|
TNRC18
|
trinucleotide repeat containing 18 |
| chr12_-_121802886 | 1.21 |
ENST00000545885.5
ENST00000542933.5 ENST00000428029.6 ENST00000541694.5 ENST00000536662.5 ENST00000535643.5 ENST00000541657.5 |
LINC01089
RHOF
|
long intergenic non-protein coding RNA 1089 ras homolog family member F, filopodia associated |
| chr17_+_44557476 | 1.21 |
ENST00000315323.5
|
FZD2
|
frizzled class receptor 2 |
| chrX_+_123963164 | 1.20 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr11_-_84923162 | 1.19 |
ENST00000524982.5
|
DLG2
|
discs large MAGUK scaffold protein 2 |
| chr8_-_92095432 | 1.17 |
ENST00000518954.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr17_-_40703744 | 1.16 |
ENST00000264651.3
|
KRT24
|
keratin 24 |
| chr14_-_53951880 | 1.14 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr18_-_55422492 | 1.13 |
ENST00000561992.5
ENST00000630712.2 |
TCF4
|
transcription factor 4 |
| chr9_+_2015969 | 1.12 |
ENST00000636903.1
ENST00000637352.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr16_+_2682515 | 1.11 |
ENST00000301738.9
ENST00000564195.1 |
KCTD5
|
potassium channel tetramerization domain containing 5 |
| chr8_+_10095704 | 1.09 |
ENST00000382490.9
|
MSRA
|
methionine sulfoxide reductase A |
| chr7_-_116030735 | 1.08 |
ENST00000393485.5
|
TFEC
|
transcription factor EC |
| chr15_-_53733103 | 1.08 |
ENST00000559418.5
|
WDR72
|
WD repeat domain 72 |
| chr6_-_10412367 | 1.06 |
ENST00000379608.9
|
TFAP2A
|
transcription factor AP-2 alpha |
| chr20_-_51802433 | 1.06 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4 |
| chr6_+_135851681 | 1.04 |
ENST00000308191.11
|
PDE7B
|
phosphodiesterase 7B |
| chr9_+_2717479 | 1.02 |
ENST00000382082.4
|
KCNV2
|
potassium voltage-gated channel modifier subfamily V member 2 |
| chr12_-_102480552 | 1.02 |
ENST00000337514.11
ENST00000307046.8 |
IGF1
|
insulin like growth factor 1 |
| chr1_+_192316992 | 1.01 |
ENST00000417209.2
|
RGS21
|
regulator of G protein signaling 21 |
| chr12_-_102480638 | 1.01 |
ENST00000392904.5
|
IGF1
|
insulin like growth factor 1 |
| chr5_+_51383394 | 1.00 |
ENST00000230658.12
|
ISL1
|
ISL LIM homeobox 1 |
| chr7_-_29195447 | 1.00 |
ENST00000437527.1
ENST00000455544.5 |
CPVL
|
carboxypeptidase vitellogenic like |
| chr10_+_112950452 | 1.00 |
ENST00000369397.8
|
TCF7L2
|
transcription factor 7 like 2 |
| chr9_-_95509241 | 1.00 |
ENST00000331920.11
|
PTCH1
|
patched 1 |
| chr18_-_55322334 | 0.99 |
ENST00000630720.3
|
TCF4
|
transcription factor 4 |
| chr6_+_68638540 | 0.99 |
ENST00000546190.5
|
ADGRB3
|
adhesion G protein-coupled receptor B3 |
| chr3_+_1093002 | 0.98 |
ENST00000446702.7
|
CNTN6
|
contactin 6 |
| chr2_-_144430934 | 0.98 |
ENST00000638087.1
ENST00000638007.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr10_+_113126670 | 0.98 |
ENST00000369389.6
|
TCF7L2
|
transcription factor 7 like 2 |
| chr14_-_89494051 | 0.97 |
ENST00000555034.5
ENST00000553904.1 |
FOXN3
|
forkhead box N3 |
| chr12_-_16277685 | 0.97 |
ENST00000344941.3
|
SLC15A5
|
solute carrier family 15 member 5 |
| chr12_-_13095628 | 0.96 |
ENST00000457134.6
ENST00000537302.5 |
GSG1
|
germ cell associated 1 |
| chr7_-_27143672 | 0.96 |
ENST00000222726.4
|
HOXA5
|
homeobox A5 |
| chr8_-_92095498 | 0.95 |
ENST00000520974.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr8_-_42377227 | 0.94 |
ENST00000220812.3
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr8_+_10095551 | 0.93 |
ENST00000522907.5
ENST00000528246.5 |
MSRA
|
methionine sulfoxide reductase A |
| chr13_-_94479671 | 0.93 |
ENST00000377028.10
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr10_+_112950240 | 0.93 |
ENST00000627217.3
ENST00000534894.5 ENST00000538897.5 ENST00000536810.5 ENST00000355995.8 ENST00000542695.5 ENST00000543371.5 |
TCF7L2
|
transcription factor 7 like 2 |
| chr1_+_168280872 | 0.93 |
ENST00000367821.8
|
TBX19
|
T-box transcription factor 19 |
| chr10_+_112950586 | 0.92 |
ENST00000355717.9
ENST00000352065.10 ENST00000369395.6 ENST00000545257.6 |
TCF7L2
|
transcription factor 7 like 2 |
| chr4_+_56216101 | 0.92 |
ENST00000504228.6
|
CRACD
|
capping protein inhibiting regulator of actin dynamics |
| chr3_-_62373538 | 0.91 |
ENST00000283268.8
|
FEZF2
|
FEZ family zinc finger 2 |
| chr5_-_1882744 | 0.91 |
ENST00000511126.1
|
IRX4
|
iroquois homeobox 4 |
| chr3_+_141387801 | 0.90 |
ENST00000514251.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr2_-_29074515 | 0.90 |
ENST00000331664.6
|
PCARE
|
photoreceptor cilium actin regulator |
| chr1_-_216423396 | 0.89 |
ENST00000366942.3
ENST00000674083.1 ENST00000307340.8 |
USH2A
|
usherin |
| chr1_+_244051275 | 0.88 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr17_-_48610971 | 0.88 |
ENST00000239165.9
|
HOXB7
|
homeobox B7 |
| chr12_-_13095798 | 0.87 |
ENST00000396302.7
|
GSG1
|
germ cell associated 1 |
| chr14_-_56810448 | 0.87 |
ENST00000339475.10
ENST00000555006.5 ENST00000672264.2 ENST00000554559.5 ENST00000555804.1 |
OTX2
|
orthodenticle homeobox 2 |
| chr18_-_55351977 | 0.87 |
ENST00000643689.1
|
TCF4
|
transcription factor 4 |
| chr8_-_92095468 | 0.87 |
ENST00000520583.5
ENST00000519061.5 |
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr2_+_17816458 | 0.87 |
ENST00000281047.4
|
MSGN1
|
mesogenin 1 |
| chr7_-_27140195 | 0.86 |
ENST00000522788.5
ENST00000317201.7 |
HOXA3
|
homeobox A3 |
| chr6_-_22297028 | 0.85 |
ENST00000306482.2
|
PRL
|
prolactin |
| chr8_+_103819212 | 0.84 |
ENST00000515551.5
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr3_+_152299392 | 0.84 |
ENST00000498502.5
ENST00000545754.5 ENST00000357472.7 ENST00000324196.9 |
MBNL1
|
muscleblind like splicing regulator 1 |
| chr9_-_124595808 | 0.83 |
ENST00000475178.1
|
NR6A1
|
nuclear receptor subfamily 6 group A member 1 |
| chr17_-_10549694 | 0.83 |
ENST00000622564.4
|
MYH2
|
myosin heavy chain 2 |
| chr2_+_44941695 | 0.83 |
ENST00000260653.5
|
SIX3
|
SIX homeobox 3 |
| chr1_+_40709475 | 0.83 |
ENST00000372651.5
|
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr11_-_33892010 | 0.82 |
ENST00000257818.3
|
LMO2
|
LIM domain only 2 |
| chr12_-_102480604 | 0.82 |
ENST00000392905.7
|
IGF1
|
insulin like growth factor 1 |
| chr17_-_48530910 | 0.79 |
ENST00000577092.1
ENST00000239174.7 |
HOXB1
|
homeobox B1 |
| chr7_-_42152396 | 0.79 |
ENST00000642432.1
ENST00000647255.1 ENST00000677288.1 |
GLI3
|
GLI family zinc finger 3 |
| chr2_+_176116768 | 0.79 |
ENST00000249501.5
|
HOXD10
|
homeobox D10 |
| chr4_-_41748713 | 0.79 |
ENST00000226382.4
|
PHOX2B
|
paired like homeobox 2B |
| chr11_+_20599602 | 0.78 |
ENST00000525748.6
|
SLC6A5
|
solute carrier family 6 member 5 |
| chr7_+_114416286 | 0.76 |
ENST00000635534.1
|
FOXP2
|
forkhead box P2 |
| chr8_-_121641424 | 0.75 |
ENST00000303924.5
|
HAS2
|
hyaluronan synthase 2 |
| chr10_-_49396074 | 0.75 |
ENST00000374139.8
|
DRGX
|
dorsal root ganglia homeobox |
| chr6_-_154247422 | 0.74 |
ENST00000519190.1
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr14_-_24081928 | 0.73 |
ENST00000396995.1
|
NRL
|
neural retina leucine zipper |
| chr14_-_24081986 | 0.73 |
ENST00000560550.1
|
NRL
|
neural retina leucine zipper |
| chr15_+_58431985 | 0.73 |
ENST00000433326.2
ENST00000299022.10 |
LIPC
|
lipase C, hepatic type |
| chr14_+_61187544 | 0.72 |
ENST00000555185.5
ENST00000557294.5 ENST00000556778.5 |
PRKCH
|
protein kinase C eta |
| chr14_-_56810380 | 0.72 |
ENST00000672125.1
ENST00000673481.1 |
OTX2
|
orthodenticle homeobox 2 |
| chr2_-_162838728 | 0.72 |
ENST00000328032.8
ENST00000332142.10 |
KCNH7
|
potassium voltage-gated channel subfamily H member 7 |
| chr7_-_151167692 | 0.72 |
ENST00000297537.5
|
GBX1
|
gastrulation brain homeobox 1 |
| chr8_-_115668609 | 0.72 |
ENST00000220888.9
|
TRPS1
|
transcriptional repressor GATA binding 1 |
| chr12_-_91004965 | 0.71 |
ENST00000261172.8
|
EPYC
|
epiphycan |
| chr4_+_87799546 | 0.71 |
ENST00000226284.7
|
IBSP
|
integrin binding sialoprotein |
| chr1_-_152159227 | 0.71 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr7_+_18495723 | 0.70 |
ENST00000681950.1
ENST00000622668.4 ENST00000405010.7 ENST00000406451.8 ENST00000441542.7 ENST00000428307.6 ENST00000681273.1 |
HDAC9
|
histone deacetylase 9 |
| chr7_-_84492718 | 0.70 |
ENST00000424555.5
|
SEMA3A
|
semaphorin 3A |
| chr3_+_107524357 | 0.70 |
ENST00000456817.5
ENST00000458458.5 |
BBX
|
BBX high mobility group box domain containing |
| chr17_-_10549652 | 0.70 |
ENST00000245503.10
|
MYH2
|
myosin heavy chain 2 |
| chr12_-_23944301 | 0.70 |
ENST00000538083.1
|
SOX5
|
SRY-box transcription factor 5 |
| chr7_+_38977904 | 0.69 |
ENST00000518318.7
ENST00000403058.6 |
POU6F2
|
POU class 6 homeobox 2 |
| chr17_-_55511434 | 0.69 |
ENST00000636752.1
|
SMIM36
|
small integral membrane protein 36 |
| chr5_-_131532993 | 0.69 |
ENST00000504575.5
ENST00000513227.5 |
RAPGEF6
|
Rap guanine nucleotide exchange factor 6 |
| chr4_+_157220795 | 0.69 |
ENST00000506284.5
|
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr18_+_75210755 | 0.68 |
ENST00000322038.5
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr2_-_144520006 | 0.68 |
ENST00000303660.8
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr16_-_67483541 | 0.68 |
ENST00000290953.3
|
AGRP
|
agouti related neuropeptide |
| chr12_-_116881062 | 0.67 |
ENST00000550505.5
|
HRK
|
harakiri, BCL2 interacting protein |
| chr16_-_1884231 | 0.65 |
ENST00000563416.3
ENST00000633813.1 ENST00000470044.5 |
LINC00254
MEIOB
|
long intergenic non-protein coding RNA 254 meiosis specific with OB-fold |
| chr17_-_10549612 | 0.65 |
ENST00000532183.6
ENST00000397183.6 ENST00000420805.1 |
MYH2
|
myosin heavy chain 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 1.6 | 8.2 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 1.4 | 4.1 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 1.2 | 5.8 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.9 | 0.9 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.9 | 13.0 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.8 | 3.3 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.8 | 3.9 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.7 | 0.7 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.7 | 2.1 | GO:0021757 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.6 | 3.2 | GO:0035910 | N-terminal peptidyl-lysine acetylation(GO:0018076) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.6 | 1.9 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.6 | 2.5 | GO:0060873 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.6 | 2.8 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.5 | 1.0 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.5 | 6.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.5 | 1.4 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.5 | 2.3 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.4 | 2.6 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.4 | 1.3 | GO:0048561 | establishment of organ orientation(GO:0048561) |
| 0.4 | 1.7 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.4 | 1.2 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) positive regulation of ovulation(GO:0060279) |
| 0.4 | 2.8 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.4 | 0.8 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.4 | 2.6 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.4 | 1.8 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.3 | 4.6 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.3 | 1.5 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.3 | 2.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.3 | 8.9 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 1.9 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.3 | 4.9 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.3 | 2.7 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.3 | 0.8 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.3 | 0.8 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.2 | 1.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.2 | 0.7 | GO:0048867 | ganglion mother cell fate determination(GO:0007402) stem cell fate determination(GO:0048867) |
| 0.2 | 1.6 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.2 | 4.2 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.2 | 4.8 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 3.9 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.2 | 2.0 | GO:0002254 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.2 | 3.0 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 1.5 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 0.8 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 | 3.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 0.9 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 1.9 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.2 | 2.4 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.2 | 1.0 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.2 | 1.6 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.2 | 0.9 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.2 | 0.8 | GO:0036233 | glycine import(GO:0036233) |
| 0.2 | 2.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 2.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 3.2 | GO:0071600 | otic vesicle morphogenesis(GO:0071600) |
| 0.1 | 2.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 4.7 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 3.8 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 1.3 | GO:0090325 | regulation of locomotion involved in locomotory behavior(GO:0090325) |
| 0.1 | 1.0 | GO:0099558 | motor learning(GO:0061743) maintenance of synapse structure(GO:0099558) |
| 0.1 | 1.9 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 2.6 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 2.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 2.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 2.2 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.6 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.4 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.7 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.7 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 0.5 | GO:0021592 | fourth ventricle development(GO:0021592) initiation of neural tube closure(GO:0021993) |
| 0.1 | 6.0 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 2.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 2.0 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 13.5 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 2.9 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 9.3 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 1.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.5 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.1 | 0.3 | GO:0060849 | radial pattern formation(GO:0009956) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 | 1.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.6 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.9 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.3 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 1.2 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 0.6 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.2 | GO:0010768 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 0.1 | 0.2 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.1 | 0.7 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 1.5 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 1.9 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.5 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.7 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 0.3 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.7 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 1.0 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 1.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.9 | GO:0090494 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 1.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.9 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 1.0 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 1.2 | GO:0072662 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 1.0 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.0 | 2.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 2.4 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 7.9 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.8 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.8 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 1.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 1.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.8 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 2.0 | GO:0045446 | endothelial cell differentiation(GO:0045446) |
| 0.0 | 0.2 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 1.2 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.2 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 1.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.5 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 1.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 2.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.0 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.1 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 13.0 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.7 | 2.0 | GO:0016590 | ACF complex(GO:0016590) |
| 0.4 | 1.2 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 0.9 | GO:0002141 | stereocilia ankle link(GO:0002141) USH2 complex(GO:1990696) |
| 0.3 | 2.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 4.8 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.3 | 2.8 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.3 | 1.9 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.3 | 5.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 2.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 2.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 5.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 5.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.3 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 1.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.0 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 1.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 2.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 2.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 13.9 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 3.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 11.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 16.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 5.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.0 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.9 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.2 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 5.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 2.5 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 2.3 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 3.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 9.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 2.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.0 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 1.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 8.2 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 1.2 | 4.7 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.5 | 6.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.4 | 4.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.4 | 1.7 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.4 | 2.2 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.3 | 2.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.3 | 8.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.3 | 1.9 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.3 | 1.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 5.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 2.7 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 0.8 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.2 | 3.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 0.8 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.2 | 0.9 | GO:0005334 | norepinephrine:sodium symporter activity(GO:0005334) |
| 0.2 | 0.9 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.2 | 2.7 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.2 | 4.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 1.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 0.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.0 | GO:0097108 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.1 | 12.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 3.9 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 3.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.8 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.7 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 3.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 6.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.6 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 4.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 10.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 1.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 7.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 5.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 2.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 2.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.7 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 2.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 2.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 2.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 2.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 1.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 2.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 1.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 2.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 46.9 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.5 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 2.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.7 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.7 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 1.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.9 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 2.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.4 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 1.8 | GO:0000975 | regulatory region DNA binding(GO:0000975) regulatory region nucleic acid binding(GO:0001067) transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 1.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 7.8 | GO:0003700 | transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 14.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 4.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 5.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 13.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 4.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 2.8 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 3.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 3.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 3.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 4.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 9.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.2 | 3.9 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 4.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 2.7 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 4.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 4.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.8 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 2.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 8.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 3.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 2.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.1 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 2.4 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |