Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for HOXB5
Z-value: 0.69
Transcription factors associated with HOXB5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB5
|
ENSG00000120075.6 | homeobox B5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB5 | hg38_v1_chr17_-_48593748_48593786 | -0.30 | 9.6e-02 | Click! |
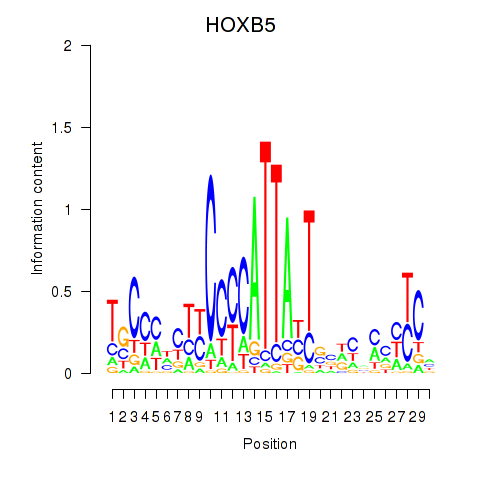
Activity profile of HOXB5 motif
Sorted Z-values of HOXB5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_161223559 | 4.59 |
ENST00000469730.2
ENST00000463273.5 ENST00000464492.5 ENST00000367990.7 ENST00000470459.6 ENST00000463812.1 ENST00000468465.5 |
APOA2
|
apolipoprotein A2 |
| chr1_-_161223408 | 3.14 |
ENST00000491350.1
|
APOA2
|
apolipoprotein A2 |
| chr7_+_5282935 | 2.77 |
ENST00000396872.8
ENST00000444741.5 ENST00000297195.8 ENST00000406453.3 |
SLC29A4
|
solute carrier family 29 member 4 |
| chr5_-_35047935 | 2.58 |
ENST00000510428.1
ENST00000231420.11 |
AGXT2
|
alanine--glyoxylate aminotransferase 2 |
| chrX_+_48521817 | 2.40 |
ENST00000446158.5
ENST00000414061.1 |
EBP
|
EBP cholestenol delta-isomerase |
| chrX_+_48521788 | 2.32 |
ENST00000651615.1
ENST00000495186.6 |
ENSG00000286268.1
EBP
|
novel protein EBP cholestenol delta-isomerase |
| chr10_-_101843765 | 2.10 |
ENST00000370046.5
|
KCNIP2
|
potassium voltage-gated channel interacting protein 2 |
| chr10_-_101843777 | 2.00 |
ENST00000356640.7
|
KCNIP2
|
potassium voltage-gated channel interacting protein 2 |
| chr15_-_74725370 | 1.75 |
ENST00000567032.5
ENST00000564596.5 ENST00000566503.1 ENST00000395049.8 ENST00000379727.8 ENST00000617691.4 ENST00000395048.6 |
CYP1A1
|
cytochrome P450 family 1 subfamily A member 1 |
| chr3_-_165196689 | 1.61 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chr3_-_127822455 | 1.56 |
ENST00000265052.10
|
MGLL
|
monoglyceride lipase |
| chr19_-_15934410 | 1.46 |
ENST00000326742.12
|
CYP4F11
|
cytochrome P450 family 4 subfamily F member 11 |
| chr19_-_15934521 | 1.19 |
ENST00000402119.9
|
CYP4F11
|
cytochrome P450 family 4 subfamily F member 11 |
| chrM_+_12329 | 1.16 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 5 |
| chr3_+_190563440 | 0.72 |
ENST00000453359.5
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr10_+_112376193 | 0.68 |
ENST00000433418.6
ENST00000356116.6 ENST00000354273.5 |
ACSL5
|
acyl-CoA synthetase long chain family member 5 |
| chr11_-_35419098 | 0.67 |
ENST00000606205.6
ENST00000645303.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr1_+_162381703 | 0.67 |
ENST00000458626.4
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr2_-_74499583 | 0.64 |
ENST00000377566.9
|
LBX2
|
ladybird homeobox 2 |
| chr11_+_63506046 | 0.63 |
ENST00000674247.1
|
LGALS12
|
galectin 12 |
| chr11_-_35418966 | 0.61 |
ENST00000531628.2
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr2_-_187565751 | 0.61 |
ENST00000421427.5
|
TFPI
|
tissue factor pathway inhibitor |
| chr2_+_17540670 | 0.60 |
ENST00000451533.5
ENST00000295156.9 |
VSNL1
|
visinin like 1 |
| chr17_-_7207340 | 0.58 |
ENST00000650301.1
|
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr11_+_109422174 | 0.55 |
ENST00000327419.7
|
C11orf87
|
chromosome 11 open reading frame 87 |
| chr4_-_151325488 | 0.55 |
ENST00000604030.7
|
SH3D19
|
SH3 domain containing 19 |
| chr1_+_235328728 | 0.55 |
ENST00000646281.1
|
TBCE
|
tubulin folding cofactor E |
| chr2_-_46941760 | 0.53 |
ENST00000444761.6
ENST00000409147.1 |
MCFD2
|
multiple coagulation factor deficiency 2, ER cargo receptor complex subunit |
| chr3_-_187702557 | 0.52 |
ENST00000358241.1
|
RTP2
|
receptor transporter protein 2 |
| chr3_+_51385282 | 0.49 |
ENST00000528157.7
|
MANF
|
mesencephalic astrocyte derived neurotrophic factor |
| chr1_+_21981099 | 0.48 |
ENST00000400277.2
|
CELA3B
|
chymotrypsin like elastase 3B |
| chr21_-_44262216 | 0.48 |
ENST00000270172.7
|
DNMT3L
|
DNA methyltransferase 3 like |
| chr9_+_4662281 | 0.48 |
ENST00000381883.5
|
PLPP6
|
phospholipid phosphatase 6 |
| chr7_+_130381092 | 0.46 |
ENST00000484324.1
|
CPA1
|
carboxypeptidase A1 |
| chr14_+_21030509 | 0.45 |
ENST00000481535.5
|
TPPP2
|
tubulin polymerization promoting protein family member 2 |
| chr16_-_57971086 | 0.44 |
ENST00000564448.5
ENST00000311183.8 |
CNGB1
|
cyclic nucleotide gated channel subunit beta 1 |
| chr2_+_17541157 | 0.44 |
ENST00000406397.1
|
VSNL1
|
visinin like 1 |
| chr17_+_48892761 | 0.43 |
ENST00000355938.9
ENST00000393366.7 ENST00000503641.5 ENST00000514808.5 ENST00000506855.1 |
ATP5MC1
|
ATP synthase membrane subunit c locus 1 |
| chr14_+_21030201 | 0.42 |
ENST00000321760.11
ENST00000460647.6 ENST00000530140.6 ENST00000472458.5 |
TPPP2
|
tubulin polymerization promoting protein family member 2 |
| chr1_+_4654732 | 0.41 |
ENST00000378190.7
|
AJAP1
|
adherens junctions associated protein 1 |
| chr11_-_57322300 | 0.40 |
ENST00000527207.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1 |
| chr16_-_57971121 | 0.39 |
ENST00000251102.13
|
CNGB1
|
cyclic nucleotide gated channel subunit beta 1 |
| chr1_+_40691998 | 0.38 |
ENST00000534399.5
ENST00000372653.5 |
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr2_-_46941710 | 0.37 |
ENST00000409207.5
|
MCFD2
|
multiple coagulation factor deficiency 2, ER cargo receptor complex subunit |
| chrX_-_107982370 | 0.36 |
ENST00000302917.1
|
TEX13B
|
testis expressed 13B |
| chr12_-_52321395 | 0.36 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr17_-_44066595 | 0.35 |
ENST00000585388.2
ENST00000293406.8 |
LSM12
|
LSM12 homolog |
| chr15_-_50765656 | 0.33 |
ENST00000261854.10
|
SPPL2A
|
signal peptide peptidase like 2A |
| chr19_-_13953302 | 0.32 |
ENST00000585607.1
ENST00000538517.6 ENST00000587458.1 ENST00000538371.6 |
PODNL1
|
podocan like 1 |
| chr8_-_92095498 | 0.31 |
ENST00000520974.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr6_-_107957252 | 0.31 |
ENST00000429168.1
|
SEC63
|
SEC63 homolog, protein translocation regulator |
| chr11_-_57322197 | 0.30 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1 |
| chr12_-_7665897 | 0.30 |
ENST00000229304.5
|
APOBEC1
|
apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| chr8_-_92095432 | 0.29 |
ENST00000518954.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr8_-_132085655 | 0.29 |
ENST00000262283.5
|
ENSG00000258417.3
|
novel protein |
| chrX_+_14529516 | 0.28 |
ENST00000218075.9
|
GLRA2
|
glycine receptor alpha 2 |
| chr3_+_45945019 | 0.27 |
ENST00000458629.1
ENST00000457814.1 |
CXCR6
|
C-X-C motif chemokine receptor 6 |
| chr8_-_92095410 | 0.26 |
ENST00000518823.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr11_-_6030758 | 0.26 |
ENST00000641900.1
|
OR56A1
|
olfactory receptor family 56 subfamily A member 1 |
| chr4_-_67754335 | 0.25 |
ENST00000420975.2
ENST00000226413.5 |
GNRHR
|
gonadotropin releasing hormone receptor |
| chr1_-_247458105 | 0.24 |
ENST00000641149.1
ENST00000641527.1 |
OR2B11
|
olfactory receptor family 2 subfamily B member 11 |
| chr1_-_155688294 | 0.24 |
ENST00000311573.9
|
YY1AP1
|
YY1 associated protein 1 |
| chr1_+_40691749 | 0.23 |
ENST00000372654.5
|
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr11_-_118225086 | 0.22 |
ENST00000640745.1
|
JAML
|
junction adhesion molecule like |
| chr1_+_198639162 | 0.21 |
ENST00000418674.1
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr19_+_7507082 | 0.19 |
ENST00000599312.1
|
ENSG00000267952.1
|
novel transcript |
| chr14_+_22105305 | 0.19 |
ENST00000390453.1
|
TRAV24
|
T cell receptor alpha variable 24 |
| chr7_+_142580911 | 0.19 |
ENST00000621184.1
|
TRBV12-5
|
T cell receptor beta variable 12-5 |
| chr2_-_219308963 | 0.18 |
ENST00000423636.6
ENST00000442029.5 ENST00000412847.5 |
PTPRN
|
protein tyrosine phosphatase receptor type N |
| chr8_-_92095468 | 0.18 |
ENST00000520583.5
ENST00000519061.5 |
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr11_+_4643966 | 0.18 |
ENST00000396952.6
|
OR51E1
|
olfactory receptor family 51 subfamily E member 1 |
| chr1_+_155610218 | 0.17 |
ENST00000649846.1
ENST00000245564.8 ENST00000368341.8 |
MSTO1
|
misato mitochondrial distribution and morphology regulator 1 |
| chr1_+_235328399 | 0.17 |
ENST00000471812.5
ENST00000358966.6 ENST00000282841.9 ENST00000391855.2 ENST00000647428.1 ENST00000645655.1 ENST00000647186.1 ENST00000646624.1 ENST00000645205.1 ENST00000645351.1 |
GGPS1
TBCE
|
geranylgeranyl diphosphate synthase 1 tubulin folding cofactor E |
| chr6_+_29459771 | 0.16 |
ENST00000377132.1
|
OR2H1
|
olfactory receptor family 2 subfamily H member 1 |
| chr19_+_13151975 | 0.16 |
ENST00000588173.1
|
IER2
|
immediate early response 2 |
| chr9_-_127937800 | 0.15 |
ENST00000373110.4
ENST00000314392.13 |
DPM2
|
dolichyl-phosphate mannosyltransferase subunit 2, regulatory |
| chr1_+_40691689 | 0.15 |
ENST00000427410.6
ENST00000447388.7 ENST00000425457.6 ENST00000453631.5 ENST00000456393.6 |
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr5_-_141682211 | 0.15 |
ENST00000239440.9
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr8_-_92095598 | 0.15 |
ENST00000520724.5
ENST00000518844.5 |
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr11_-_56748696 | 0.15 |
ENST00000641581.1
ENST00000641668.1 |
OR9G4
|
olfactory receptor family 9 subfamily G member 4 |
| chr1_-_36385872 | 0.14 |
ENST00000373129.7
|
STK40
|
serine/threonine kinase 40 |
| chr5_-_141682192 | 0.14 |
ENST00000508305.5
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr12_-_6689359 | 0.14 |
ENST00000683879.1
|
ZNF384
|
zinc finger protein 384 |
| chr3_-_195811889 | 0.13 |
ENST00000475231.5
|
MUC4
|
mucin 4, cell surface associated |
| chr6_+_26021641 | 0.13 |
ENST00000617569.2
|
H4C1
|
H4 clustered histone 1 |
| chr3_-_195811916 | 0.12 |
ENST00000463781.8
|
MUC4
|
mucin 4, cell surface associated |
| chr1_-_36385887 | 0.11 |
ENST00000373130.7
ENST00000373132.4 |
STK40
|
serine/threonine kinase 40 |
| chr5_-_161546970 | 0.11 |
ENST00000675303.1
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr8_-_33597750 | 0.09 |
ENST00000522982.1
|
DUSP26
|
dual specificity phosphatase 26 |
| chr5_+_150190035 | 0.09 |
ENST00000230671.7
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 member 7 |
| chr5_-_172283743 | 0.08 |
ENST00000393792.3
|
UBTD2
|
ubiquitin domain containing 2 |
| chr11_-_128522189 | 0.06 |
ENST00000526145.6
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr20_-_50691536 | 0.06 |
ENST00000327979.8
|
RIPOR3
|
RIPOR family member 3 |
| chr8_+_117135259 | 0.04 |
ENST00000519688.5
|
SLC30A8
|
solute carrier family 30 member 8 |
| chr15_+_41621134 | 0.04 |
ENST00000566718.6
|
MGA
|
MAX dimerization protein MGA |
| chr1_-_155688727 | 0.02 |
ENST00000404643.5
ENST00000359205.9 ENST00000407221.5 ENST00000355499.9 |
YY1AP1
|
YY1 associated protein 1 |
| chr9_-_110208156 | 0.02 |
ENST00000400613.5
|
C9orf152
|
chromosome 9 open reading frame 152 |
| chr15_+_41621492 | 0.01 |
ENST00000570161.6
|
MGA
|
MAX dimerization protein MGA |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.7 | GO:2000910 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.8 | 4.7 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.7 | 2.6 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.6 | 2.6 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.6 | 1.7 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.4 | 4.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 1.6 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 2.8 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 0.3 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.3 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) regulation of mRNA modification(GO:0090365) |
| 0.1 | 0.7 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 1.3 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.4 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.2 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.1 | 0.6 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.8 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.3 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.4 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.0 | 0.2 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.0 | 0.5 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.2 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 1.0 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.7 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 1.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.7 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 1.2 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 0.8 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.4 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.3 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 1.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 4.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.2 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) cholestenol delta-isomerase activity(GO:0047750) |
| 1.5 | 7.7 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.5 | 4.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.4 | 2.6 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.3 | 1.7 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.2 | 2.8 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.2 | 0.6 | GO:0030395 | lactose binding(GO:0030395) |
| 0.1 | 0.8 | GO:0005223 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.7 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 1.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 1.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.3 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 2.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.3 | GO:0019958 | C-X-C chemokine receptor activity(GO:0016494) C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.7 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 4.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 2.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.6 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 1.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |