Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
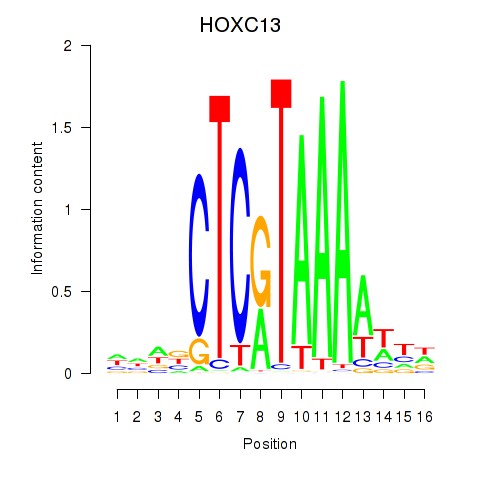
Results for HOXC13
Z-value: 0.76
Transcription factors associated with HOXC13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC13
|
ENSG00000123364.5 | homeobox C13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC13 | hg38_v1_chr12_+_53938824_53938852 | 0.21 | 2.5e-01 | Click! |
Activity profile of HOXC13 motif
Sorted Z-values of HOXC13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_70666961 | 5.49 |
ENST00000510437.5
|
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr4_-_70666884 | 3.36 |
ENST00000510614.5
|
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr10_-_46046264 | 2.80 |
ENST00000581478.5
ENST00000582163.3 |
MSMB
|
microseminoprotein beta |
| chr6_+_87407965 | 2.36 |
ENST00000369562.9
|
CFAP206
|
cilia and flagella associated protein 206 |
| chr8_-_85378105 | 2.25 |
ENST00000521846.5
ENST00000523022.6 ENST00000524324.5 ENST00000519991.5 ENST00000520663.5 ENST00000517590.5 ENST00000522579.5 ENST00000522814.5 ENST00000522662.5 ENST00000523858.5 ENST00000519129.5 |
CA1
|
carbonic anhydrase 1 |
| chr3_-_149377637 | 2.24 |
ENST00000305366.8
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr4_-_154612635 | 1.97 |
ENST00000407946.5
ENST00000405164.5 ENST00000336098.8 ENST00000393846.6 ENST00000404648.7 ENST00000443553.5 |
FGG
|
fibrinogen gamma chain |
| chr1_+_192575765 | 1.76 |
ENST00000469578.2
ENST00000367459.8 |
RGS1
|
regulator of G protein signaling 1 |
| chr7_-_16833411 | 1.67 |
ENST00000412973.1
|
AGR2
|
anterior gradient 2, protein disulphide isomerase family member |
| chr6_-_52803807 | 1.65 |
ENST00000334575.6
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr18_-_63644250 | 1.64 |
ENST00000341074.10
ENST00000436264.1 |
SERPINB4
|
serpin family B member 4 |
| chr6_-_49713564 | 1.59 |
ENST00000616725.4
ENST00000618917.4 |
CRISP2
|
cysteine rich secretory protein 2 |
| chr7_-_41703062 | 1.56 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr8_-_27992663 | 1.56 |
ENST00000380385.6
ENST00000354914.8 |
SCARA5
|
scavenger receptor class A member 5 |
| chr6_-_49713521 | 1.47 |
ENST00000339139.5
|
CRISP2
|
cysteine rich secretory protein 2 |
| chr3_-_123620496 | 1.46 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr22_+_22758698 | 1.45 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr4_+_20251896 | 1.39 |
ENST00000504154.6
|
SLIT2
|
slit guidance ligand 2 |
| chr2_-_189179754 | 1.38 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr12_-_91146195 | 1.37 |
ENST00000548218.1
|
DCN
|
decorin |
| chr14_-_106335613 | 1.36 |
ENST00000603660.1
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr6_+_127577168 | 1.33 |
ENST00000329722.8
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr6_-_52763473 | 1.33 |
ENST00000493422.3
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr3_-_172523460 | 1.33 |
ENST00000420541.6
|
TNFSF10
|
TNF superfamily member 10 |
| chr1_-_109113818 | 1.32 |
ENST00000369949.8
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr3_-_123620571 | 1.31 |
ENST00000583087.5
|
MYLK
|
myosin light chain kinase |
| chr2_+_33134620 | 1.29 |
ENST00000402934.5
ENST00000404525.5 ENST00000407925.5 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr3_-_172523423 | 1.28 |
ENST00000241261.7
|
TNFSF10
|
TNF superfamily member 10 |
| chr1_-_209802149 | 1.20 |
ENST00000456314.1
|
IRF6
|
interferon regulatory factor 6 |
| chr4_+_168711416 | 1.18 |
ENST00000649826.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr14_-_106507476 | 1.17 |
ENST00000390621.3
|
IGHV1-45
|
immunoglobulin heavy variable 1-45 |
| chr9_-_92404690 | 1.17 |
ENST00000447356.1
|
OGN
|
osteoglycin |
| chr2_+_33134579 | 1.16 |
ENST00000418533.6
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr17_-_76643748 | 1.10 |
ENST00000156626.12
|
ST6GALNAC1
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 |
| chr9_-_92482499 | 1.04 |
ENST00000375544.7
|
ASPN
|
asporin |
| chr15_+_70936487 | 1.04 |
ENST00000558456.5
ENST00000560158.6 ENST00000558808.5 ENST00000559806.5 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr8_-_94262308 | 1.04 |
ENST00000297596.3
ENST00000396194.6 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr3_+_130581741 | 1.03 |
ENST00000511332.1
|
COL6A6
|
collagen type VI alpha 6 chain |
| chr18_-_27143024 | 1.00 |
ENST00000581714.5
|
CHST9
|
carbohydrate sulfotransferase 9 |
| chr22_+_22322452 | 0.99 |
ENST00000390290.3
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr1_+_43300971 | 0.99 |
ENST00000372476.8
ENST00000538015.1 |
TIE1
|
tyrosine kinase with immunoglobulin like and EGF like domains 1 |
| chr7_+_134866831 | 0.90 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr9_-_92482350 | 0.90 |
ENST00000375543.2
|
ASPN
|
asporin |
| chr8_-_27992624 | 0.88 |
ENST00000524352.5
|
SCARA5
|
scavenger receptor class A member 5 |
| chr9_+_110125491 | 0.88 |
ENST00000259318.7
|
PALM2AKAP2
|
PALM2 and AKAP2 fusion |
| chr1_+_109114097 | 0.88 |
ENST00000457623.6
ENST00000369939.8 ENST00000529753.5 |
ELAPOR1
|
endosome-lysosome associated apoptosis and autophagy regulator 1 |
| chr3_+_111911604 | 0.86 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology like domain family B member 2 |
| chr3_-_100993409 | 0.84 |
ENST00000471714.6
|
ABI3BP
|
ABI family member 3 binding protein |
| chr12_-_91153149 | 0.78 |
ENST00000550758.1
|
DCN
|
decorin |
| chr12_-_114403898 | 0.78 |
ENST00000526441.1
|
TBX5
|
T-box transcription factor 5 |
| chr20_-_45791865 | 0.78 |
ENST00000243938.9
|
WFDC3
|
WAP four-disulfide core domain 3 |
| chr2_+_102337148 | 0.77 |
ENST00000311734.6
ENST00000409584.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr20_+_38317627 | 0.76 |
ENST00000417318.3
|
BPI
|
bactericidal permeability increasing protein |
| chr5_-_180649579 | 0.74 |
ENST00000261937.11
ENST00000502649.5 |
FLT4
|
fms related receptor tyrosine kinase 4 |
| chr15_+_57599411 | 0.74 |
ENST00000569089.1
|
MYZAP
|
myocardial zonula adherens protein |
| chr1_-_169711603 | 0.74 |
ENST00000236147.6
ENST00000650983.1 |
SELL
|
selectin L |
| chr2_+_106063234 | 0.73 |
ENST00000409944.5
|
ECRG4
|
ECRG4 augurin precursor |
| chr2_+_191245185 | 0.73 |
ENST00000418908.5
ENST00000339514.8 ENST00000392318.8 |
MYO1B
|
myosin IB |
| chr11_+_118304721 | 0.72 |
ENST00000361763.9
|
CD3E
|
CD3e molecule |
| chr15_+_70892443 | 0.72 |
ENST00000443425.6
ENST00000560369.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr9_+_98943705 | 0.71 |
ENST00000610452.1
|
COL15A1
|
collagen type XV alpha 1 chain |
| chr3_-_99850976 | 0.69 |
ENST00000487087.5
|
FILIP1L
|
filamin A interacting protein 1 like |
| chr1_+_174875505 | 0.69 |
ENST00000486220.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr6_-_117425905 | 0.68 |
ENST00000368507.8
|
ROS1
|
ROS proto-oncogene 1, receptor tyrosine kinase |
| chr12_+_47079586 | 0.67 |
ENST00000432328.2
ENST00000546455.6 |
PCED1B
|
PC-esterase domain containing 1B |
| chr9_-_92482461 | 0.65 |
ENST00000651738.1
|
ASPN
|
asporin |
| chr1_-_155910881 | 0.65 |
ENST00000609492.1
ENST00000368322.7 |
RIT1
|
Ras like without CAAX 1 |
| chr3_-_51875597 | 0.65 |
ENST00000446461.2
|
IQCF5
|
IQ motif containing F5 |
| chr1_-_89126066 | 0.64 |
ENST00000370466.4
|
GBP2
|
guanylate binding protein 2 |
| chr5_+_42756811 | 0.64 |
ENST00000388827.4
ENST00000361970.10 |
CCDC152
|
coiled-coil domain containing 152 |
| chr10_-_20897288 | 0.62 |
ENST00000377122.9
|
NEBL
|
nebulette |
| chr19_+_53866139 | 0.60 |
ENST00000421337.6
ENST00000651087.1 |
MYADM
|
myeloid associated differentiation marker |
| chr12_-_52814106 | 0.60 |
ENST00000551956.2
|
KRT4
|
keratin 4 |
| chr19_-_18522051 | 0.59 |
ENST00000262809.9
|
ELL
|
elongation factor for RNA polymerase II |
| chr6_+_87472925 | 0.59 |
ENST00000369556.7
ENST00000369557.9 ENST00000369552.9 |
SLC35A1
|
solute carrier family 35 member A1 |
| chr1_-_178869272 | 0.59 |
ENST00000444255.1
|
ANGPTL1
|
angiopoietin like 1 |
| chr7_+_23710203 | 0.58 |
ENST00000422637.5
ENST00000355870.8 |
STK31
|
serine/threonine kinase 31 |
| chr14_-_91867529 | 0.57 |
ENST00000435962.7
|
TC2N
|
tandem C2 domains, nuclear |
| chr6_-_144008118 | 0.57 |
ENST00000629195.2
ENST00000650125.1 ENST00000647880.1 ENST00000649307.1 ENST00000674357.1 ENST00000417959.4 ENST00000635591.1 |
PLAGL1
HYMAI
|
PLAG1 like zinc finger 1 hydatidiform mole associated and imprinted |
| chr4_-_174283614 | 0.57 |
ENST00000513696.1
ENST00000393674.7 ENST00000503293.5 |
FBXO8
|
F-box protein 8 |
| chr2_+_137964446 | 0.56 |
ENST00000280096.5
ENST00000280097.5 |
HNMT
|
histamine N-methyltransferase |
| chr15_-_58279245 | 0.54 |
ENST00000558231.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr7_+_23710326 | 0.54 |
ENST00000354639.7
ENST00000531170.5 ENST00000444333.2 |
STK31
|
serine/threonine kinase 31 |
| chr14_+_49598910 | 0.54 |
ENST00000298288.11
|
LRR1
|
leucine rich repeat protein 1 |
| chr18_+_63887698 | 0.54 |
ENST00000457692.5
ENST00000299502.9 ENST00000413956.5 |
SERPINB2
|
serpin family B member 2 |
| chr6_+_30489501 | 0.53 |
ENST00000376630.5
|
HLA-E
|
major histocompatibility complex, class I, E |
| chr6_-_46954922 | 0.53 |
ENST00000265417.7
|
ADGRF5
|
adhesion G protein-coupled receptor F5 |
| chr3_-_160399207 | 0.52 |
ENST00000465537.5
ENST00000486856.5 ENST00000468218.5 ENST00000478370.5 ENST00000326448.12 |
IFT80
|
intraflagellar transport 80 |
| chr5_+_111073309 | 0.52 |
ENST00000379706.4
|
TSLP
|
thymic stromal lymphopoietin |
| chr11_+_101914997 | 0.52 |
ENST00000263468.13
|
CEP126
|
centrosomal protein 126 |
| chr6_+_33272067 | 0.51 |
ENST00000474973.5
ENST00000439602.7 |
RPS18
|
ribosomal protein S18 |
| chr6_+_42155399 | 0.50 |
ENST00000623004.2
ENST00000372963.4 ENST00000654459.1 |
GUCA1ANB
GUCA1A
|
GUCA1A neighbor guanylate cyclase activator 1A |
| chr1_-_149861210 | 0.50 |
ENST00000579512.2
|
H4C15
|
H4 clustered histone 15 |
| chr7_-_29112894 | 0.50 |
ENST00000448959.5
|
CPVL
|
carboxypeptidase vitellogenic like |
| chr1_+_173635332 | 0.49 |
ENST00000417563.3
|
TEX50
|
testis expressed 50 |
| chr3_-_160399247 | 0.49 |
ENST00000489004.5
ENST00000496589.5 |
IFT80
|
intraflagellar transport 80 |
| chr9_-_127771335 | 0.49 |
ENST00000373276.7
ENST00000373277.8 |
SH2D3C
|
SH2 domain containing 3C |
| chr10_+_50990864 | 0.49 |
ENST00000401604.8
|
PRKG1
|
protein kinase cGMP-dependent 1 |
| chr6_-_73523757 | 0.49 |
ENST00000455918.2
ENST00000677236.1 |
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr3_+_50173807 | 0.49 |
ENST00000420831.1
|
SEMA3F
|
semaphorin 3F |
| chr7_+_23710263 | 0.49 |
ENST00000433467.6
|
STK31
|
serine/threonine kinase 31 |
| chr17_+_50373214 | 0.48 |
ENST00000393271.6
ENST00000338165.9 ENST00000511519.6 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr9_-_128656649 | 0.48 |
ENST00000372715.7
|
DYNC2I2
|
dynein 2 intermediate chain 2 |
| chr2_+_33134406 | 0.48 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr16_-_67999468 | 0.48 |
ENST00000393847.6
ENST00000573808.1 ENST00000572624.5 |
DPEP2
|
dipeptidase 2 |
| chr11_+_102112445 | 0.48 |
ENST00000524575.5
|
YAP1
|
Yes1 associated transcriptional regulator |
| chr6_-_36547400 | 0.47 |
ENST00000229812.8
|
STK38
|
serine/threonine kinase 38 |
| chr19_+_8842593 | 0.47 |
ENST00000305625.2
|
MBD3L1
|
methyl-CpG binding domain protein 3 like 1 |
| chr6_-_32371872 | 0.47 |
ENST00000527965.5
ENST00000532023.5 ENST00000447241.6 ENST00000534588.1 |
TSBP1
|
testis expressed basic protein 1 |
| chr4_-_163473732 | 0.46 |
ENST00000280605.5
|
TKTL2
|
transketolase like 2 |
| chr15_-_99249523 | 0.46 |
ENST00000560235.1
ENST00000394132.7 ENST00000560860.5 ENST00000558078.5 ENST00000560772.5 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr6_+_133953210 | 0.45 |
ENST00000367869.1
ENST00000237264.9 ENST00000674115.1 |
TBPL1
ENSG00000288529.1
|
TATA-box binding protein like 1 novel protein |
| chr5_-_180649613 | 0.45 |
ENST00000393347.7
ENST00000619105.4 |
FLT4
|
fms related receptor tyrosine kinase 4 |
| chr4_+_89901979 | 0.45 |
ENST00000508372.1
|
MMRN1
|
multimerin 1 |
| chr13_-_102798958 | 0.45 |
ENST00000376004.5
|
POGLUT2
|
protein O-glucosyltransferase 2 |
| chr11_+_85855377 | 0.44 |
ENST00000342404.8
|
CCDC83
|
coiled-coil domain containing 83 |
| chr12_-_9208388 | 0.44 |
ENST00000261336.7
|
PZP
|
PZP alpha-2-macroglobulin like |
| chr14_-_67412112 | 0.44 |
ENST00000216446.9
|
PLEK2
|
pleckstrin 2 |
| chr6_-_32371897 | 0.44 |
ENST00000442822.6
ENST00000375015.8 ENST00000533191.5 |
TSBP1
|
testis expressed basic protein 1 |
| chr3_+_57890011 | 0.43 |
ENST00000494088.6
ENST00000438794.5 |
SLMAP
|
sarcolemma associated protein |
| chr10_+_100997040 | 0.42 |
ENST00000370223.7
|
LZTS2
|
leucine zipper tumor suppressor 2 |
| chr9_-_2844058 | 0.42 |
ENST00000397885.3
|
PUM3
|
pumilio RNA binding family member 3 |
| chr3_+_42936354 | 0.40 |
ENST00000383748.9
|
KRBOX1
|
KRAB box domain containing 1 |
| chr1_+_54641806 | 0.39 |
ENST00000409996.5
|
MROH7
|
maestro heat like repeat family member 7 |
| chr17_-_76643648 | 0.39 |
ENST00000589992.1
|
ST6GALNAC1
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 |
| chr19_+_44905785 | 0.39 |
ENST00000446996.5
ENST00000252486.9 ENST00000434152.5 |
APOE
|
apolipoprotein E |
| chr1_+_54641754 | 0.38 |
ENST00000339553.9
ENST00000421030.7 |
MROH7
|
maestro heat like repeat family member 7 |
| chr1_+_206406377 | 0.38 |
ENST00000605476.5
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr1_+_158845798 | 0.37 |
ENST00000438394.1
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr1_-_46176482 | 0.36 |
ENST00000540385.2
ENST00000506599.2 |
P3R3URF-PIK3R3
P3R3URF
|
P3R3URF-PIK3R3 readthrough PIK3R3 upstream reading frame |
| chr1_+_159009886 | 0.36 |
ENST00000340979.10
ENST00000368131.8 ENST00000295809.12 ENST00000368132.7 |
IFI16
|
interferon gamma inducible protein 16 |
| chr12_-_57237090 | 0.36 |
ENST00000556732.1
|
NDUFA4L2
|
NDUFA4 mitochondrial complex associated like 2 |
| chr12_-_54295748 | 0.35 |
ENST00000540264.2
ENST00000312156.8 |
NFE2
|
nuclear factor, erythroid 2 |
| chr12_-_13095628 | 0.35 |
ENST00000457134.6
ENST00000537302.5 |
GSG1
|
germ cell associated 1 |
| chr6_-_75243770 | 0.35 |
ENST00000472311.6
ENST00000460985.1 ENST00000377978.3 ENST00000684430.1 ENST00000509698.6 |
COX7A2
|
cytochrome c oxidase subunit 7A2 |
| chr12_+_96489569 | 0.35 |
ENST00000524981.9
|
CFAP54
|
cilia and flagella associated protein 54 |
| chr19_+_16888991 | 0.34 |
ENST00000248076.4
|
F2RL3
|
F2R like thrombin or trypsin receptor 3 |
| chr1_+_39215255 | 0.33 |
ENST00000671089.1
|
MACF1
|
microtubule actin crosslinking factor 1 |
| chr19_-_6737565 | 0.33 |
ENST00000601716.1
ENST00000264080.11 |
GPR108
|
G protein-coupled receptor 108 |
| chr20_+_38962299 | 0.33 |
ENST00000373325.6
ENST00000373323.8 ENST00000252011.8 ENST00000615559.1 |
DHX35
ENSG00000277581.1
|
DEAH-box helicase 35 novel transcript, sense intronic to DHX35 |
| chr5_+_35852695 | 0.33 |
ENST00000508941.5
|
IL7R
|
interleukin 7 receptor |
| chr1_-_79188467 | 0.33 |
ENST00000656300.1
|
ADGRL4
|
adhesion G protein-coupled receptor L4 |
| chr19_-_35228699 | 0.32 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187 member B |
| chr1_+_149832647 | 0.32 |
ENST00000578186.2
|
H4C14
|
H4 clustered histone 14 |
| chr4_-_185471082 | 0.32 |
ENST00000507501.5
ENST00000506962.3 |
CCDC110
|
coiled-coil domain containing 110 |
| chr16_-_11273610 | 0.31 |
ENST00000327157.4
|
PRM3
|
protamine 3 |
| chr17_-_58692032 | 0.31 |
ENST00000349033.10
ENST00000389934.7 |
TEX14
|
testis expressed 14, intercellular bridge forming factor |
| chr4_+_69995958 | 0.31 |
ENST00000381060.2
ENST00000246895.9 |
STATH
|
statherin |
| chr7_-_25180278 | 0.31 |
ENST00000283905.8
ENST00000409280.5 ENST00000415598.5 |
C7orf31
|
chromosome 7 open reading frame 31 |
| chr7_+_23680130 | 0.30 |
ENST00000409192.7
ENST00000409653.5 ENST00000409994.3 ENST00000344962.9 |
FAM221A
|
family with sequence similarity 221 member A |
| chr14_+_96256194 | 0.30 |
ENST00000216629.11
ENST00000553356.1 |
BDKRB1
|
bradykinin receptor B1 |
| chr3_-_108953870 | 0.29 |
ENST00000261047.8
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr19_+_3762665 | 0.29 |
ENST00000330133.5
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr3_-_108953762 | 0.29 |
ENST00000393963.7
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr22_+_32043253 | 0.29 |
ENST00000266088.9
|
SLC5A1
|
solute carrier family 5 member 1 |
| chr10_+_30434602 | 0.29 |
ENST00000413724.5
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr17_-_8383164 | 0.29 |
ENST00000584164.6
ENST00000582556.5 ENST00000648839.1 ENST00000578812.5 ENST00000583011.6 |
RPL26
|
ribosomal protein L26 |
| chr2_+_161136901 | 0.28 |
ENST00000259075.6
ENST00000432002.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr22_+_39520747 | 0.28 |
ENST00000676430.1
|
ATF4
|
activating transcription factor 4 |
| chr2_+_102418642 | 0.28 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr7_-_93148345 | 0.28 |
ENST00000437805.5
ENST00000446959.5 ENST00000439952.5 ENST00000414791.5 ENST00000446033.1 ENST00000411955.5 ENST00000318238.9 |
SAMD9L
|
sterile alpha motif domain containing 9 like |
| chrX_+_56729231 | 0.28 |
ENST00000637096.1
ENST00000374922.9 ENST00000423617.2 |
NBDY
|
negative regulator of P-body association |
| chr22_-_39532722 | 0.28 |
ENST00000334678.8
|
RPS19BP1
|
ribosomal protein S19 binding protein 1 |
| chr4_-_1208825 | 0.27 |
ENST00000511679.5
ENST00000617421.4 |
SPON2
|
spondin 2 |
| chr21_+_38256698 | 0.26 |
ENST00000613499.4
ENST00000612702.4 ENST00000398925.5 ENST00000398928.5 ENST00000328656.8 ENST00000443341.5 |
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr9_+_121121760 | 0.25 |
ENST00000373847.5
|
CNTRL
|
centriolin |
| chr1_+_54641816 | 0.25 |
ENST00000395690.6
|
MROH7
|
maestro heat like repeat family member 7 |
| chr7_+_23710236 | 0.25 |
ENST00000456014.6
|
STK31
|
serine/threonine kinase 31 |
| chr6_-_32371912 | 0.25 |
ENST00000612031.4
|
TSBP1
|
testis expressed basic protein 1 |
| chr6_-_32816910 | 0.25 |
ENST00000447394.1
ENST00000438763.7 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr3_+_129440196 | 0.25 |
ENST00000507564.5
ENST00000348417.7 ENST00000504021.5 ENST00000349441.6 |
IFT122
|
intraflagellar transport 122 |
| chr13_+_33818122 | 0.24 |
ENST00000380071.8
|
RFC3
|
replication factor C subunit 3 |
| chr2_+_161160420 | 0.24 |
ENST00000392749.7
ENST00000405852.5 |
TANK
|
TRAF family member associated NFKB activator |
| chrX_+_13689171 | 0.24 |
ENST00000618931.2
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr11_-_114559866 | 0.24 |
ENST00000534921.2
|
NXPE1
|
neurexophilin and PC-esterase domain family member 1 |
| chr16_+_11965193 | 0.24 |
ENST00000053243.6
ENST00000396495.3 |
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr4_-_149815826 | 0.22 |
ENST00000636793.2
ENST00000636414.1 |
IQCM
|
IQ motif containing M |
| chr3_+_178419123 | 0.21 |
ENST00000614557.1
ENST00000455307.5 ENST00000436432.1 |
ENSG00000275163.1
LINC01014
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 (KCNMB2-IT1 - KCNMB2 readthrough transcript) long intergenic non-protein coding RNA 1014 |
| chr10_-_77637902 | 0.21 |
ENST00000286627.10
ENST00000639486.1 ENST00000640523.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr22_+_39520553 | 0.21 |
ENST00000674920.3
ENST00000679776.1 ENST00000675582.2 ENST00000337304.2 ENST00000676346.2 ENST00000396680.3 ENST00000680446.1 ENST00000674568.2 ENST00000680748.1 ENST00000674835.2 |
ATF4
|
activating transcription factor 4 |
| chr6_+_133953156 | 0.20 |
ENST00000367871.5
|
TBPL1
|
TATA-box binding protein like 1 |
| chr11_-_114559847 | 0.19 |
ENST00000251921.6
|
NXPE1
|
neurexophilin and PC-esterase domain family member 1 |
| chr3_-_179451387 | 0.19 |
ENST00000675901.1
ENST00000232564.8 ENST00000674862.1 ENST00000497513.1 |
GNB4
|
G protein subunit beta 4 |
| chr12_-_15662692 | 0.19 |
ENST00000540613.5
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr5_+_150640652 | 0.19 |
ENST00000307662.5
|
SYNPO
|
synaptopodin |
| chr17_-_58692021 | 0.19 |
ENST00000240361.12
|
TEX14
|
testis expressed 14, intercellular bridge forming factor |
| chr22_-_37844308 | 0.18 |
ENST00000411961.6
ENST00000434930.1 ENST00000215941.9 |
ANKRD54
|
ankyrin repeat domain 54 |
| chrX_+_13689116 | 0.18 |
ENST00000464506.2
ENST00000243325.6 |
RAB9A
|
RAB9A, member RAS oncogene family |
| chr21_+_46286623 | 0.18 |
ENST00000397691.1
|
YBEY
|
ybeY metalloendoribonuclease |
| chr9_-_34329205 | 0.18 |
ENST00000684219.1
ENST00000402558.7 |
KIF24
|
kinesin family member 24 |
| chr4_+_53377749 | 0.18 |
ENST00000507166.5
|
ENSG00000282278.1
|
novel FIP1L1-PDGFRA fusion protein |
| chr19_-_56393430 | 0.18 |
ENST00000589895.1
ENST00000589143.5 ENST00000301310.8 ENST00000586929.5 |
ZNF582
|
zinc finger protein 582 |
| chr9_-_72060605 | 0.18 |
ENST00000377024.8
ENST00000651200.2 ENST00000652752.1 |
C9orf57
|
chromosome 9 open reading frame 57 |
| chr6_+_144330797 | 0.17 |
ENST00000628146.2
|
UTRN
|
utrophin |
| chr6_+_43130583 | 0.16 |
ENST00000481946.5
|
PTK7
|
protein tyrosine kinase 7 (inactive) |
| chr19_-_8698705 | 0.16 |
ENST00000612068.1
|
ACTL9
|
actin like 9 |
| chr6_+_150721073 | 0.16 |
ENST00000358517.6
|
PLEKHG1
|
pleckstrin homology and RhoGEF domain containing G1 |
| chr6_-_11807045 | 0.15 |
ENST00000379415.6
|
ADTRP
|
androgen dependent TFPI regulating protein |
| chr12_+_92702983 | 0.15 |
ENST00000344636.6
ENST00000544406.2 |
PLEKHG7
|
pleckstrin homology and RhoGEF domain containing G7 |
| chr7_-_86965872 | 0.15 |
ENST00000398276.6
ENST00000416314.5 ENST00000425689.1 |
ELAPOR2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 8.9 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.7 | 2.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.5 | 1.6 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.5 | 1.4 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.3 | 1.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 | 1.7 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.3 | 2.9 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 2.6 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.3 | 2.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 0.8 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.3 | 1.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 1.8 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 0.7 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.4 | GO:0010877 | lipid transport involved in lipid storage(GO:0010877) regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.5 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.5 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.7 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 2.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.7 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 2.6 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 0.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 3.0 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 0.6 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.6 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 1.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 2.5 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.1 | 0.4 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.3 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.1 | 2.4 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 0.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 0.5 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 1.2 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 0.7 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.1 | 1.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 1.3 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 1.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.4 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) |
| 0.1 | 0.2 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.3 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.5 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.0 | 0.5 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.3 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.6 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 1.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.4 | GO:0052405 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 2.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.4 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 2.7 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.5 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 1.2 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 4.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 1.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.5 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 1.0 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.3 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.5 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.1 | GO:0045355 | negative regulation of interferon-alpha biosynthetic process(GO:0045355) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.0 | 1.7 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.5 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.4 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.9 | GO:0071757 | hexameric IgM immunoglobulin complex(GO:0071757) |
| 0.8 | 2.4 | GO:0001534 | radial spoke(GO:0001534) |
| 0.5 | 1.6 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 1.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 2.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 0.5 | GO:1990617 | CHOP-ATF4 complex(GO:1990617) |
| 0.2 | 2.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.6 | GO:0035363 | histone locus body(GO:0035363) |
| 0.1 | 2.9 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.4 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.5 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 0.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.7 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.1 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 1.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 2.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 1.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0042611 | MHC protein complex(GO:0042611) MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 5.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 8.9 | GO:0019862 | IgA binding(GO:0019862) |
| 0.5 | 2.9 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.4 | 1.2 | GO:0036328 | VEGF-C-activated receptor activity(GO:0036328) |
| 0.3 | 0.8 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.3 | 2.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 2.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 1.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 1.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 1.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 1.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.7 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.3 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.3 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 0.5 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 0.7 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.4 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 1.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 2.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 1.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 2.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.5 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 5.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 1.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.0 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 2.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 5.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 1.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 3.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 2.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 4.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 2.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 3.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 3.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 2.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |