Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
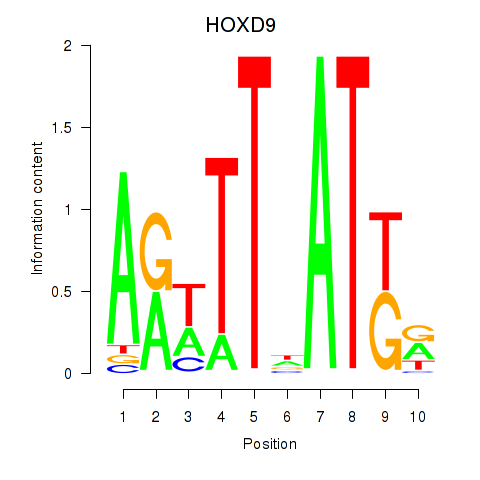
Results for HOXD9
Z-value: 0.92
Transcription factors associated with HOXD9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD9
|
ENSG00000128709.13 | homeobox D9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD9 | hg38_v1_chr2_+_176122712_176122727 | -0.19 | 3.0e-01 | Click! |
Activity profile of HOXD9 motif
Sorted Z-values of HOXD9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_10849464 | 3.28 |
ENST00000544994.5
ENST00000228811.8 ENST00000540107.2 |
PRR4
|
proline rich 4 |
| chr2_-_85668172 | 2.56 |
ENST00000428225.5
ENST00000519937.7 |
SFTPB
|
surfactant protein B |
| chr4_-_76023489 | 2.43 |
ENST00000306602.3
|
CXCL10
|
C-X-C motif chemokine ligand 10 |
| chr12_-_91153149 | 2.28 |
ENST00000550758.1
|
DCN
|
decorin |
| chr3_-_116444983 | 2.21 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr1_+_81306096 | 1.90 |
ENST00000370721.5
ENST00000370727.5 ENST00000370725.5 ENST00000370723.5 ENST00000370728.5 ENST00000370730.5 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr5_+_95391361 | 1.85 |
ENST00000283357.10
|
FAM81B
|
family with sequence similarity 81 member B |
| chr7_-_16881967 | 1.60 |
ENST00000402239.7
ENST00000310398.7 ENST00000414935.1 |
AGR3
|
anterior gradient 3, protein disulphide isomerase family member |
| chr4_+_168712159 | 1.59 |
ENST00000510998.5
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr3_-_139539577 | 1.27 |
ENST00000619087.4
|
RBP1
|
retinol binding protein 1 |
| chr3_-_139539679 | 1.23 |
ENST00000483943.6
ENST00000672186.1 ENST00000232219.6 ENST00000617459.4 ENST00000492918.1 |
RBP1
|
retinol binding protein 1 |
| chr3_-_149377637 | 1.22 |
ENST00000305366.8
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr17_+_9576627 | 1.20 |
ENST00000396219.7
ENST00000352665.10 |
CFAP52
|
cilia and flagella associated protein 52 |
| chr17_+_70075215 | 1.17 |
ENST00000283936.5
ENST00000615244.4 ENST00000392671.6 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr6_+_118548289 | 1.15 |
ENST00000357525.6
|
PLN
|
phospholamban |
| chr3_-_100846405 | 1.15 |
ENST00000495591.5
ENST00000466947.5 |
ABI3BP
|
ABI family member 3 binding protein |
| chr1_+_37556913 | 1.13 |
ENST00000296218.8
ENST00000652629.1 |
DNALI1
|
dynein axonemal light intermediate chain 1 |
| chr1_+_15756659 | 1.11 |
ENST00000375771.5
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr4_-_76007501 | 1.10 |
ENST00000264888.6
|
CXCL9
|
C-X-C motif chemokine ligand 9 |
| chr1_+_160400543 | 1.09 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr1_-_94925759 | 1.07 |
ENST00000415017.1
ENST00000545882.5 |
CNN3
|
calponin 3 |
| chr8_-_85341659 | 1.04 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr22_+_22906342 | 1.04 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr16_-_3372666 | 0.96 |
ENST00000399974.5
|
MTRNR2L4
|
MT-RNR2 like 4 |
| chr9_-_92482350 | 0.95 |
ENST00000375543.2
|
ASPN
|
asporin |
| chr1_-_58577244 | 0.92 |
ENST00000371225.4
|
TACSTD2
|
tumor associated calcium signal transducer 2 |
| chr4_+_73740541 | 0.92 |
ENST00000401931.1
ENST00000307407.8 |
CXCL8
|
C-X-C motif chemokine ligand 8 |
| chr6_-_110358313 | 0.90 |
ENST00000338882.5
|
METTL24
|
methyltransferase like 24 |
| chr9_+_12693327 | 0.90 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
| chr6_+_52423680 | 0.88 |
ENST00000538167.2
|
EFHC1
|
EF-hand domain containing 1 |
| chr12_-_88580459 | 0.87 |
ENST00000552044.1
ENST00000644744.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr22_-_30246739 | 0.85 |
ENST00000403987.3
ENST00000249075.4 |
LIF
|
LIF interleukin 6 family cytokine |
| chr6_-_49713521 | 0.84 |
ENST00000339139.5
|
CRISP2
|
cysteine rich secretory protein 2 |
| chr5_+_122129533 | 0.83 |
ENST00000296600.5
ENST00000504912.1 ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chr13_+_75839716 | 0.82 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr3_+_194136138 | 0.79 |
ENST00000232424.4
|
HES1
|
hes family bHLH transcription factor 1 |
| chr12_-_120325936 | 0.78 |
ENST00000549767.1
|
PLA2G1B
|
phospholipase A2 group IB |
| chr2_+_89947508 | 0.78 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr4_+_168711416 | 0.73 |
ENST00000649826.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr4_-_167234426 | 0.72 |
ENST00000541354.5
ENST00000509854.5 ENST00000512681.5 ENST00000357545.9 ENST00000510741.5 ENST00000510403.5 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr1_+_15756603 | 0.72 |
ENST00000496928.6
ENST00000508310.5 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr1_+_15756628 | 0.68 |
ENST00000510393.5
ENST00000430076.5 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr2_-_187448244 | 0.68 |
ENST00000392370.8
ENST00000410068.5 ENST00000447403.5 ENST00000410102.5 |
CALCRL
|
calcitonin receptor like receptor |
| chr13_-_44474250 | 0.67 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family member 1 |
| chr12_-_9116223 | 0.67 |
ENST00000404455.2
|
A2M
|
alpha-2-macroglobulin |
| chr3_+_178558700 | 0.66 |
ENST00000432997.5
ENST00000455865.5 |
KCNMB2
|
potassium calcium-activated channel subfamily M regulatory beta subunit 2 |
| chr17_+_70075317 | 0.66 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr1_+_81699665 | 0.64 |
ENST00000359929.7
|
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr20_+_59835853 | 0.63 |
ENST00000492611.5
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr1_-_25905098 | 0.62 |
ENST00000374291.5
|
STMN1
|
stathmin 1 |
| chr7_+_118224654 | 0.61 |
ENST00000265224.9
ENST00000486422.1 ENST00000417525.5 |
ANKRD7
|
ankyrin repeat domain 7 |
| chr6_-_106975616 | 0.61 |
ENST00000610952.1
|
CD24
|
CD24 molecule |
| chr3_-_167474026 | 0.60 |
ENST00000466903.1
ENST00000264677.8 |
SERPINI2
|
serpin family I member 2 |
| chr1_+_166989089 | 0.60 |
ENST00000367870.6
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr19_-_56314788 | 0.60 |
ENST00000592509.5
ENST00000592679.5 ENST00000683990.1 ENST00000588442.5 ENST00000593106.5 ENST00000587492.5 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr10_+_113751675 | 0.60 |
ENST00000369312.9
ENST00000619563.5 |
PLEKHS1
|
pleckstrin homology domain containing S1 |
| chr11_-_102530738 | 0.58 |
ENST00000260227.5
|
MMP7
|
matrix metallopeptidase 7 |
| chr3_-_146528750 | 0.58 |
ENST00000483300.5
|
PLSCR1
|
phospholipid scramblase 1 |
| chr8_-_85341705 | 0.55 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr1_+_100719734 | 0.55 |
ENST00000370119.8
ENST00000294728.7 ENST00000347652.6 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr5_-_79512794 | 0.55 |
ENST00000282260.10
ENST00000508576.5 ENST00000535690.1 |
HOMER1
|
homer scaffold protein 1 |
| chr2_-_174847525 | 0.55 |
ENST00000295497.12
ENST00000652036.1 ENST00000444394.6 ENST00000650731.1 |
CHN1
|
chimerin 1 |
| chr5_-_139439488 | 0.55 |
ENST00000302060.10
|
DNAJC18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr3_-_167653916 | 0.55 |
ENST00000488012.5
ENST00000682715.1 ENST00000647816.1 |
WDR49
|
WD repeat domain 49 |
| chr2_+_209580024 | 0.54 |
ENST00000392194.5
|
MAP2
|
microtubule associated protein 2 |
| chr4_-_167234579 | 0.53 |
ENST00000502330.5
ENST00000357154.7 ENST00000421836.6 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr11_+_112176364 | 0.53 |
ENST00000526088.5
ENST00000532593.5 ENST00000531169.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr10_+_5364955 | 0.52 |
ENST00000380433.5
|
UCN3
|
urocortin 3 |
| chr5_+_141208697 | 0.51 |
ENST00000624949.1
ENST00000622978.1 ENST00000239450.4 |
PCDHB12
|
protocadherin beta 12 |
| chr6_-_49866527 | 0.50 |
ENST00000335847.9
|
CRISP1
|
cysteine rich secretory protein 1 |
| chr12_+_19205294 | 0.48 |
ENST00000424268.5
|
PLEKHA5
|
pleckstrin homology domain containing A5 |
| chr1_+_44674688 | 0.47 |
ENST00000418644.5
ENST00000458657.6 ENST00000535358.6 ENST00000441519.5 ENST00000445071.5 |
ARMH1
|
armadillo like helical domain containing 1 |
| chr12_-_30735014 | 0.46 |
ENST00000433722.6
|
CAPRIN2
|
caprin family member 2 |
| chr12_-_56741535 | 0.46 |
ENST00000647707.1
|
ENSG00000285625.1
|
novel protein |
| chr9_-_121050264 | 0.45 |
ENST00000223642.3
|
C5
|
complement C5 |
| chr22_-_35840218 | 0.45 |
ENST00000414461.6
ENST00000416721.6 ENST00000449924.6 ENST00000262829.11 ENST00000397305.3 |
RBFOX2
|
RNA binding fox-1 homolog 2 |
| chr6_-_49866453 | 0.45 |
ENST00000507853.5
|
CRISP1
|
cysteine rich secretory protein 1 |
| chr22_-_41286168 | 0.44 |
ENST00000356244.8
|
RANGAP1
|
Ran GTPase activating protein 1 |
| chr18_+_616672 | 0.44 |
ENST00000338387.11
|
CLUL1
|
clusterin like 1 |
| chr4_-_185649524 | 0.43 |
ENST00000451974.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr10_+_102854251 | 0.43 |
ENST00000339834.10
ENST00000369883.3 |
BORCS7
|
BLOC-1 related complex subunit 7 |
| chr12_+_40742342 | 0.43 |
ENST00000548005.5
ENST00000552248.5 |
CNTN1
|
contactin 1 |
| chr13_-_33350619 | 0.42 |
ENST00000439831.1
ENST00000567873.1 |
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr2_-_174847765 | 0.42 |
ENST00000443238.6
|
CHN1
|
chimerin 1 |
| chr10_+_52128343 | 0.42 |
ENST00000672084.1
|
PRKG1
|
protein kinase cGMP-dependent 1 |
| chr11_+_67586104 | 0.42 |
ENST00000495996.1
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr4_-_69639642 | 0.41 |
ENST00000604629.6
ENST00000604021.1 |
UGT2A2
|
UDP glucuronosyltransferase family 2 member A2 |
| chr11_+_112175526 | 0.41 |
ENST00000532612.5
ENST00000438022.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr14_+_61697622 | 0.41 |
ENST00000539097.2
|
HIF1A
|
hypoxia inducible factor 1 subunit alpha |
| chr3_+_2892199 | 0.41 |
ENST00000397459.6
|
CNTN4
|
contactin 4 |
| chr3_-_99850976 | 0.40 |
ENST00000487087.5
|
FILIP1L
|
filamin A interacting protein 1 like |
| chr12_+_119334696 | 0.39 |
ENST00000536742.5
|
CCDC60
|
coiled-coil domain containing 60 |
| chr6_-_131000722 | 0.39 |
ENST00000528282.5
|
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr8_+_54616057 | 0.39 |
ENST00000637698.1
|
RP1
|
RP1 axonemal microtubule associated |
| chr11_-_13495984 | 0.39 |
ENST00000282091.6
|
PTH
|
parathyroid hormone |
| chr11_-_13496018 | 0.38 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr4_+_70195719 | 0.38 |
ENST00000683306.1
|
ODAM
|
odontogenic, ameloblast associated |
| chr7_-_122699108 | 0.37 |
ENST00000340112.3
|
RNF133
|
ring finger protein 133 |
| chr15_-_99249523 | 0.37 |
ENST00000560235.1
ENST00000394132.7 ENST00000560860.5 ENST00000558078.5 ENST00000560772.5 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr18_+_616711 | 0.36 |
ENST00000579494.1
|
CLUL1
|
clusterin like 1 |
| chr8_+_78516329 | 0.36 |
ENST00000396418.7
ENST00000352966.9 |
PKIA
|
cAMP-dependent protein kinase inhibitor alpha |
| chr3_-_27456743 | 0.35 |
ENST00000295736.9
ENST00000428386.5 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4 member 7 |
| chr2_+_172821575 | 0.35 |
ENST00000397087.7
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr17_-_73227700 | 0.34 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104 member A |
| chr12_+_28452493 | 0.34 |
ENST00000542801.5
|
CCDC91
|
coiled-coil domain containing 91 |
| chr3_-_167653952 | 0.33 |
ENST00000466760.5
ENST00000479765.5 |
WDR49
|
WD repeat domain 49 |
| chr17_+_56153458 | 0.32 |
ENST00000318698.6
ENST00000682825.1 ENST00000566473.6 |
ANKFN1
|
ankyrin repeat and fibronectin type III domain containing 1 |
| chr12_+_55681711 | 0.31 |
ENST00000394252.4
|
METTL7B
|
methyltransferase like 7B |
| chr17_-_64263221 | 0.31 |
ENST00000258991.7
ENST00000583738.1 ENST00000584379.6 |
TEX2
|
testis expressed 2 |
| chr4_-_144019287 | 0.31 |
ENST00000638448.1
ENST00000513128.5 ENST00000506516.6 ENST00000429670.3 ENST00000502664.6 |
GYPB
|
glycophorin B (MNS blood group) |
| chr4_+_15339818 | 0.31 |
ENST00000397700.6
ENST00000295297.4 |
C1QTNF7
|
C1q and TNF related 7 |
| chr2_+_11534039 | 0.30 |
ENST00000381486.7
|
GREB1
|
growth regulating estrogen receptor binding 1 |
| chrX_-_131289266 | 0.30 |
ENST00000370910.5
ENST00000370901.4 ENST00000361420.8 ENST00000370903.8 |
IGSF1
|
immunoglobulin superfamily member 1 |
| chr3_+_159069252 | 0.30 |
ENST00000640015.1
ENST00000476809.7 ENST00000485419.7 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr8_-_25424260 | 0.30 |
ENST00000421054.7
|
GNRH1
|
gonadotropin releasing hormone 1 |
| chr12_-_95116967 | 0.30 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr4_+_70383123 | 0.29 |
ENST00000304915.8
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chr7_+_90383672 | 0.29 |
ENST00000416322.5
|
CLDN12
|
claudin 12 |
| chr5_+_69565122 | 0.29 |
ENST00000507595.1
|
GTF2H2C
|
GTF2H2 family member C |
| chr3_-_157503339 | 0.29 |
ENST00000392833.6
|
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr19_-_49896868 | 0.28 |
ENST00000593956.5
ENST00000391826.7 |
IL4I1
|
interleukin 4 induced 1 |
| chr5_+_141199555 | 0.28 |
ENST00000624887.1
ENST00000354757.5 |
PCDHB11
|
protocadherin beta 11 |
| chr8_+_78591222 | 0.28 |
ENST00000518467.1
|
PKIA
|
cAMP-dependent protein kinase inhibitor alpha |
| chr6_-_39431313 | 0.28 |
ENST00000229913.9
ENST00000394362.5 |
KIF6
|
kinesin family member 6 |
| chr3_-_157160751 | 0.27 |
ENST00000461804.5
|
CCNL1
|
cyclin L1 |
| chr1_-_48472166 | 0.27 |
ENST00000371847.8
ENST00000396199.7 |
SPATA6
|
spermatogenesis associated 6 |
| chr6_-_69699124 | 0.27 |
ENST00000651675.1
|
LMBRD1
|
LMBR1 domain containing 1 |
| chr15_+_58138368 | 0.27 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chr4_-_69495897 | 0.26 |
ENST00000305107.7
ENST00000639621.1 |
UGT2B4
|
UDP glucuronosyltransferase family 2 member B4 |
| chr11_-_105035113 | 0.26 |
ENST00000526568.5
ENST00000531166.5 ENST00000534497.5 ENST00000527979.5 ENST00000533400.6 ENST00000528974.1 ENST00000525825.5 ENST00000353247.9 ENST00000446369.5 ENST00000436863.7 |
CASP1
|
caspase 1 |
| chr11_+_117203135 | 0.26 |
ENST00000529622.1
|
TAGLN
|
transgelin |
| chr3_+_107599309 | 0.25 |
ENST00000406780.5
|
BBX
|
BBX high mobility group box domain containing |
| chr22_+_50170720 | 0.25 |
ENST00000159647.9
ENST00000395842.3 |
PANX2
|
pannexin 2 |
| chr1_+_86547070 | 0.24 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr5_+_141364153 | 0.24 |
ENST00000518069.2
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr9_-_28670285 | 0.24 |
ENST00000379992.6
ENST00000308675.5 ENST00000613945.3 |
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr20_+_6007245 | 0.24 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr16_+_21612637 | 0.24 |
ENST00000568826.1
|
METTL9
|
methyltransferase like 9 |
| chrX_-_77895546 | 0.23 |
ENST00000358075.11
|
MAGT1
|
magnesium transporter 1 |
| chr3_-_157503574 | 0.23 |
ENST00000494677.5
ENST00000468233.5 |
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr9_-_69014090 | 0.23 |
ENST00000377276.5
|
PRKACG
|
protein kinase cAMP-activated catalytic subunit gamma |
| chr2_-_55419821 | 0.23 |
ENST00000644630.1
ENST00000471947.2 ENST00000436346.7 ENST00000642200.1 ENST00000413716.7 ENST00000263630.13 ENST00000645072.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr2_+_233760265 | 0.22 |
ENST00000305208.10
ENST00000360418.4 |
UGT1A1
|
UDP glucuronosyltransferase family 1 member A1 |
| chr1_+_207325629 | 0.22 |
ENST00000618707.2
|
CD55
|
CD55 molecule (Cromer blood group) |
| chr4_-_83114715 | 0.22 |
ENST00000426923.2
ENST00000311507.9 ENST00000509973.5 |
PLAC8
|
placenta associated 8 |
| chr19_-_19628512 | 0.22 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr15_-_55270383 | 0.21 |
ENST00000396307.6
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr4_+_174918400 | 0.21 |
ENST00000404450.8
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr4_-_139084191 | 0.21 |
ENST00000512627.1
|
ELF2
|
E74 like ETS transcription factor 2 |
| chr10_+_84425148 | 0.21 |
ENST00000493409.5
|
CCSER2
|
coiled-coil serine rich protein 2 |
| chr1_+_173635332 | 0.21 |
ENST00000417563.3
|
TEX50
|
testis expressed 50 |
| chrX_-_77895414 | 0.20 |
ENST00000618282.5
ENST00000373336.3 |
MAGT1
|
magnesium transporter 1 |
| chr3_+_159273235 | 0.20 |
ENST00000638749.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr15_-_45201094 | 0.20 |
ENST00000561278.1
ENST00000290894.12 |
SHF
|
Src homology 2 domain containing F |
| chr3_+_141262614 | 0.20 |
ENST00000504264.5
|
PXYLP1
|
2-phosphoxylose phosphatase 1 |
| chr21_-_29061351 | 0.20 |
ENST00000432178.5
|
CCT8
|
chaperonin containing TCP1 subunit 8 |
| chr6_+_131637296 | 0.20 |
ENST00000358229.6
ENST00000357639.8 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr2_-_55296361 | 0.20 |
ENST00000647547.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr15_-_19988117 | 0.20 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr2_+_100974849 | 0.19 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr9_+_20927765 | 0.19 |
ENST00000603044.5
ENST00000604254.5 |
FOCAD
|
focadhesin |
| chr5_+_75653161 | 0.19 |
ENST00000672390.1
|
ANKDD1B
|
ankyrin repeat and death domain containing 1B |
| chr10_+_113751627 | 0.19 |
ENST00000652564.1
|
PLEKHS1
|
pleckstrin homology domain containing S1 |
| chr1_+_192158448 | 0.19 |
ENST00000367460.4
|
RGS18
|
regulator of G protein signaling 18 |
| chr14_+_93918889 | 0.19 |
ENST00000557719.1
ENST00000267594.5 |
FAM181A
|
family with sequence similarity 181 member A |
| chr14_+_37795308 | 0.19 |
ENST00000267368.11
ENST00000382320.4 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr6_-_130890393 | 0.19 |
ENST00000456097.6
|
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr5_+_126631680 | 0.19 |
ENST00000357147.4
|
TEX43
|
testis expressed 43 |
| chrX_+_48761743 | 0.19 |
ENST00000303227.11
|
GLOD5
|
glyoxalase domain containing 5 |
| chr8_-_42541042 | 0.19 |
ENST00000518717.1
|
SLC20A2
|
solute carrier family 20 member 2 |
| chr3_-_108058361 | 0.18 |
ENST00000398258.7
|
CD47
|
CD47 molecule |
| chrX_-_111270474 | 0.18 |
ENST00000324068.2
|
CAPN6
|
calpain 6 |
| chr20_-_31390580 | 0.18 |
ENST00000339144.3
ENST00000376321.4 |
DEFB119
|
defensin beta 119 |
| chr9_+_133376334 | 0.18 |
ENST00000371957.4
|
STKLD1
|
serine/threonine kinase like domain containing 1 |
| chr22_+_19479457 | 0.18 |
ENST00000407835.6
ENST00000455750.6 |
CDC45
|
cell division cycle 45 |
| chr4_-_36243939 | 0.18 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr4_+_74308463 | 0.18 |
ENST00000413830.6
|
EPGN
|
epithelial mitogen |
| chr4_-_69495861 | 0.18 |
ENST00000512583.5
|
UGT2B4
|
UDP glucuronosyltransferase family 2 member B4 |
| chr5_+_142907573 | 0.17 |
ENST00000469131.6
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr11_-_30992665 | 0.17 |
ENST00000406071.6
|
DCDC1
|
doublecortin domain containing 1 |
| chr8_+_36784324 | 0.17 |
ENST00000523973.5
ENST00000399881.8 |
KCNU1
|
potassium calcium-activated channel subfamily U member 1 |
| chr12_+_51424802 | 0.16 |
ENST00000453097.7
|
SLC4A8
|
solute carrier family 4 member 8 |
| chr6_+_10528326 | 0.16 |
ENST00000379597.7
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr14_+_23240346 | 0.16 |
ENST00000430154.6
|
RNF212B
|
ring finger protein 212B |
| chr11_+_118530990 | 0.16 |
ENST00000411589.6
ENST00000359862.8 ENST00000442938.6 |
TMEM25
|
transmembrane protein 25 |
| chr3_+_107645922 | 0.16 |
ENST00000449271.5
ENST00000425868.5 ENST00000449213.5 |
BBX
|
BBX high mobility group box domain containing |
| chr1_-_212847649 | 0.15 |
ENST00000332912.3
|
SPATA45
|
spermatogenesis associated 45 |
| chr2_+_1414382 | 0.15 |
ENST00000423320.5
ENST00000346956.7 ENST00000382198.5 |
TPO
|
thyroid peroxidase |
| chr14_-_24442662 | 0.15 |
ENST00000554698.5
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U member 1 |
| chr19_-_50639827 | 0.15 |
ENST00000593901.5
ENST00000600079.6 |
SYT3
|
synaptotagmin 3 |
| chr17_-_42112674 | 0.15 |
ENST00000251642.8
ENST00000591220.5 |
DHX58
|
DExH-box helicase 58 |
| chr20_-_53995929 | 0.15 |
ENST00000422805.1
|
BCAS1
|
brain enriched myelin associated protein 1 |
| chr1_+_198638968 | 0.15 |
ENST00000348564.11
ENST00000530727.5 ENST00000442510.8 ENST00000645247.1 ENST00000367367.8 ENST00000367364.5 ENST00000413409.6 |
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr14_+_80955043 | 0.15 |
ENST00000541158.6
|
TSHR
|
thyroid stimulating hormone receptor |
| chr18_+_58196736 | 0.15 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chrX_-_139708190 | 0.14 |
ENST00000414978.5
ENST00000519895.5 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr2_+_143129379 | 0.14 |
ENST00000295095.11
|
ARHGAP15
|
Rho GTPase activating protein 15 |
| chr16_-_21431553 | 0.14 |
ENST00000534903.1
|
NPIPB3
|
nuclear pore complex interacting protein family member B3 |
| chr5_+_141121793 | 0.14 |
ENST00000194152.4
|
PCDHB4
|
protocadherin beta 4 |
| chr19_+_15082211 | 0.14 |
ENST00000641398.1
|
OR1I1
|
olfactory receptor family 1 subfamily I member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1901876 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.3 | 0.9 | GO:0016108 | tetraterpenoid metabolic process(GO:0016108) carotenoid metabolic process(GO:0016116) carotene catabolic process(GO:0016121) xanthophyll metabolic process(GO:0016122) terpene catabolic process(GO:0046247) |
| 0.3 | 0.9 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.3 | 0.9 | GO:0070662 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) mast cell proliferation(GO:0070662) |
| 0.3 | 1.1 | GO:0060448 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.3 | 0.8 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.2 | 3.5 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.2 | 0.9 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.2 | 0.7 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 | 0.8 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.2 | 0.8 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.2 | 3.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 0.6 | GO:0032595 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.1 | 0.4 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.1 | 0.4 | GO:0035732 | nitric oxide storage(GO:0035732) |
| 0.1 | 2.5 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 2.5 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.9 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.5 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.9 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.1 | 0.4 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.1 | 0.3 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.9 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 2.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.5 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 0.5 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.3 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.4 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.1 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.4 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.1 | 2.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.6 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.2 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.2 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.1 | 0.7 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 1.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.0 | 0.4 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.6 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.3 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:1904588 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.0 | 0.2 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.4 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 3.1 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.0 | 2.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 2.5 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0075732 | viral penetration into host nucleus(GO:0075732) multi-organism nuclear import(GO:1902594) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.6 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 1.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.1 | GO:2000473 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.0 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 1.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.5 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.3 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.2 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.2 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 0.6 | GO:0030849 | autosome(GO:0030849) |
| 0.2 | 0.5 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.2 | 2.6 | GO:0097486 | alveolar lamellar body(GO:0097208) multivesicular body lumen(GO:0097486) |
| 0.2 | 2.3 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.1 | 0.4 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 0.3 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.2 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.3 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.1 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 1.2 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.8 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 2.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 2.4 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 3.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.5 | 2.5 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.2 | 0.9 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.2 | 1.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 0.9 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 0.7 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.2 | 1.6 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.4 | GO:0035730 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 0.1 | 0.5 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 2.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.3 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 0.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.7 | GO:0019959 | interleukin-8 binding(GO:0019959) tumor necrosis factor binding(GO:0043120) |
| 0.1 | 2.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.4 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.1 | 0.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 1.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 2.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 1.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.3 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.4 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0031775 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 1.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.8 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 4.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 3.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 2.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 3.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |