Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
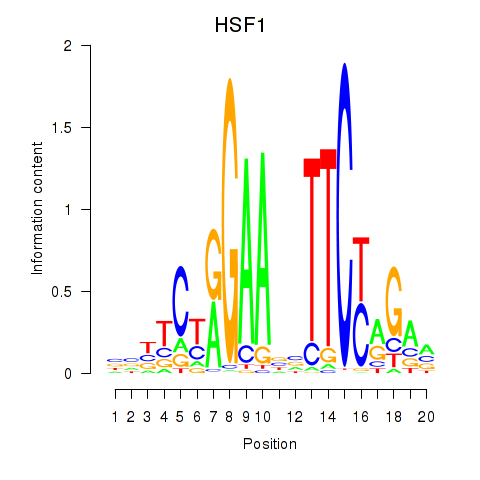
Results for HSF1
Z-value: 1.25
Transcription factors associated with HSF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF1
|
ENSG00000185122.11 | heat shock transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF1 | hg38_v1_chr8_+_144291581_144291617 | 0.47 | 7.3e-03 | Click! |
Activity profile of HSF1 motif
Sorted Z-values of HSF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_161524539 | 4.74 |
ENST00000309758.6
|
HSPA6
|
heat shock protein family A (Hsp70) member 6 |
| chrX_+_147981324 | 2.94 |
ENST00000370467.8
|
FMR1NB
|
FMR1 neighbor |
| chr3_-_46464868 | 2.79 |
ENST00000417439.5
ENST00000231751.9 ENST00000431944.1 |
LTF
|
lactotransferrin |
| chr19_+_859654 | 2.78 |
ENST00000592860.2
ENST00000327726.11 |
CFD
|
complement factor D |
| chr6_+_35297809 | 2.77 |
ENST00000316637.7
|
DEF6
|
DEF6 guanine nucleotide exchange factor |
| chr6_-_31815244 | 2.71 |
ENST00000375654.5
|
HSPA1L
|
heat shock protein family A (Hsp70) member 1 like |
| chr7_+_80646436 | 2.61 |
ENST00000419819.2
|
CD36
|
CD36 molecule |
| chr15_+_75198866 | 2.57 |
ENST00000562637.1
ENST00000360639.6 |
C15orf39
|
chromosome 15 open reading frame 39 |
| chr2_-_89213917 | 2.53 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr1_-_183569186 | 2.53 |
ENST00000420553.5
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr11_+_1870871 | 2.43 |
ENST00000417766.5
|
LSP1
|
lymphocyte specific protein 1 |
| chr5_+_119354771 | 2.42 |
ENST00000503646.1
|
TNFAIP8
|
TNF alpha induced protein 8 |
| chr3_+_114294020 | 2.40 |
ENST00000383671.8
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr20_-_45827297 | 2.39 |
ENST00000372555.8
|
TNNC2
|
troponin C2, fast skeletal type |
| chr7_+_150567382 | 2.36 |
ENST00000255945.4
ENST00000479232.1 |
GIMAP4
|
GTPase, IMAP family member 4 |
| chr7_+_150567347 | 2.35 |
ENST00000461940.5
|
GIMAP4
|
GTPase, IMAP family member 4 |
| chr7_+_80646305 | 2.25 |
ENST00000426978.5
ENST00000432207.5 |
CD36
|
CD36 molecule |
| chr2_-_89177160 | 2.16 |
ENST00000484817.1
|
IGKV2-24
|
immunoglobulin kappa variable 2-24 |
| chr20_+_45174894 | 2.16 |
ENST00000243924.4
|
PI3
|
peptidase inhibitor 3 |
| chr3_+_48223479 | 2.13 |
ENST00000652295.2
|
CAMP
|
cathelicidin antimicrobial peptide |
| chr7_+_80646347 | 2.09 |
ENST00000413265.5
|
CD36
|
CD36 molecule |
| chr6_+_31815532 | 2.09 |
ENST00000375651.7
ENST00000608703.1 |
HSPA1A
|
heat shock protein family A (Hsp70) member 1A |
| chr7_+_150737382 | 2.08 |
ENST00000358647.5
|
GIMAP5
|
GTPase, IMAP family member 5 |
| chr14_-_106335613 | 2.06 |
ENST00000603660.1
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr21_-_42315336 | 2.03 |
ENST00000398431.2
ENST00000518498.3 |
TFF3
|
trefoil factor 3 |
| chr11_-_105035113 | 1.98 |
ENST00000526568.5
ENST00000531166.5 ENST00000534497.5 ENST00000527979.5 ENST00000533400.6 ENST00000528974.1 ENST00000525825.5 ENST00000353247.9 ENST00000446369.5 ENST00000436863.7 |
CASP1
|
caspase 1 |
| chr18_+_59899988 | 1.94 |
ENST00000316660.7
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr1_-_171652675 | 1.93 |
ENST00000037502.11
|
MYOC
|
myocilin |
| chr19_-_2085324 | 1.89 |
ENST00000591638.1
|
MOB3A
|
MOB kinase activator 3A |
| chr5_+_170232912 | 1.88 |
ENST00000524171.5
ENST00000517575.4 ENST00000518395.2 ENST00000593851.5 |
C5orf58
|
chromosome 5 open reading frame 58 |
| chr11_-_19201976 | 1.83 |
ENST00000647990.1
ENST00000649235.1 ENST00000265968.9 ENST00000649842.1 |
CSRP3
|
cysteine and glycine rich protein 3 |
| chr14_-_24576240 | 1.80 |
ENST00000216336.3
|
CTSG
|
cathepsin G |
| chrX_+_52495791 | 1.78 |
ENST00000375602.2
ENST00000375600.5 |
XAGE1A
|
X antigen family member 1A |
| chr2_-_191847068 | 1.76 |
ENST00000304141.5
|
CAVIN2
|
caveolae associated protein 2 |
| chr3_+_32391841 | 1.75 |
ENST00000334983.10
|
CMTM7
|
CKLF like MARVEL transmembrane domain containing 7 |
| chr2_+_28496054 | 1.75 |
ENST00000327757.10
ENST00000422425.6 ENST00000404858.5 |
PLB1
|
phospholipase B1 |
| chr20_+_38304149 | 1.73 |
ENST00000262865.9
ENST00000642449.2 |
BPI
|
bactericidal permeability increasing protein |
| chr4_+_101790717 | 1.72 |
ENST00000508653.5
ENST00000322953.9 |
BANK1
|
B cell scaffold protein with ankyrin repeats 1 |
| chr12_+_10307950 | 1.72 |
ENST00000543420.5
ENST00000543777.5 |
KLRD1
|
killer cell lectin like receptor D1 |
| chr19_-_55325316 | 1.71 |
ENST00000591570.5
ENST00000326652.9 |
TMEM150B
|
transmembrane protein 150B |
| chr1_+_159302321 | 1.68 |
ENST00000368114.1
|
FCER1A
|
Fc fragment of IgE receptor Ia |
| chr1_-_153390976 | 1.65 |
ENST00000368732.5
ENST00000368733.4 |
S100A8
|
S100 calcium binding protein A8 |
| chr11_+_60378524 | 1.63 |
ENST00000530614.5
ENST00000530027.5 ENST00000300184.8 ENST00000530234.2 ENST00000528215.1 ENST00000531787.5 |
MS4A7
MS4A14
|
membrane spanning 4-domains A7 membrane spanning 4-domains A14 |
| chr17_+_47323941 | 1.62 |
ENST00000331493.7
ENST00000519772.5 ENST00000517484.5 |
EFCAB13
|
EF-hand calcium binding domain 13 |
| chr6_+_31827730 | 1.61 |
ENST00000375650.5
|
HSPA1B
|
heat shock protein family A (Hsp70) member 1B |
| chr17_-_48430205 | 1.60 |
ENST00000336915.11
ENST00000584924.5 |
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr5_-_172188185 | 1.60 |
ENST00000176763.10
|
STK10
|
serine/threonine kinase 10 |
| chr16_+_28931942 | 1.58 |
ENST00000324662.8
ENST00000538922.8 |
CD19
|
CD19 molecule |
| chr17_-_41505597 | 1.52 |
ENST00000336861.7
ENST00000246635.8 ENST00000587544.5 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr9_+_89311187 | 1.52 |
ENST00000314355.7
|
CKS2
|
CDC28 protein kinase regulatory subunit 2 |
| chr22_-_36160773 | 1.51 |
ENST00000531095.1
ENST00000349314.6 ENST00000397293.6 |
APOL3
|
apolipoprotein L3 |
| chr19_-_2051224 | 1.49 |
ENST00000309340.11
ENST00000589534.2 ENST00000250896.9 ENST00000589509.5 |
MKNK2
|
MAPK interacting serine/threonine kinase 2 |
| chr15_-_60397964 | 1.49 |
ENST00000558998.5
ENST00000560165.5 ENST00000557986.5 ENST00000559467.5 ENST00000677968.1 ENST00000678450.1 ENST00000332680.8 ENST00000396024.7 ENST00000557906.6 ENST00000558558.6 ENST00000559113.6 ENST00000559780.6 ENST00000559956.6 ENST00000560468.6 ENST00000678870.1 ENST00000678061.1 ENST00000451270.7 ENST00000421017.6 ENST00000560466.5 ENST00000558132.5 ENST00000559370.5 ENST00000559725.5 ENST00000558985.6 ENST00000679109.1 |
ANXA2
|
annexin A2 |
| chr4_-_83109843 | 1.47 |
ENST00000411416.6
|
PLAC8
|
placenta associated 8 |
| chr17_+_42853232 | 1.46 |
ENST00000617500.4
|
AOC3
|
amine oxidase copper containing 3 |
| chr1_+_110873135 | 1.45 |
ENST00000271324.6
|
CD53
|
CD53 molecule |
| chr17_+_42853265 | 1.44 |
ENST00000592999.5
|
AOC3
|
amine oxidase copper containing 3 |
| chr19_-_7702124 | 1.42 |
ENST00000597921.6
|
FCER2
|
Fc fragment of IgE receptor II |
| chr2_+_27051637 | 1.40 |
ENST00000451003.5
ENST00000360131.5 ENST00000323064.12 |
AGBL5
|
ATP/GTP binding protein like 5 |
| chrX_+_30215551 | 1.40 |
ENST00000378988.5
|
MAGEB2
|
MAGE family member B2 |
| chr14_-_106235582 | 1.39 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr5_+_150778733 | 1.37 |
ENST00000526627.2
|
SMIM3
|
small integral membrane protein 3 |
| chr16_+_27066919 | 1.36 |
ENST00000505035.3
|
C16orf82
|
chromosome 16 open reading frame 82 |
| chr20_+_18813777 | 1.35 |
ENST00000377428.4
|
SCP2D1
|
SCP2 sterol binding domain containing 1 |
| chr4_-_25863537 | 1.34 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr2_+_90004792 | 1.34 |
ENST00000462693.1
|
IGKV2D-24
|
immunoglobulin kappa variable 2D-24 (non-functional) |
| chr2_+_85695368 | 1.33 |
ENST00000526018.1
|
GNLY
|
granulysin |
| chrX_-_52517057 | 1.33 |
ENST00000375613.7
|
XAGE1B
|
X antigen family member 1B |
| chr11_-_26572102 | 1.32 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated |
| chr9_-_136748515 | 1.32 |
ENST00000341206.9
|
LCN6
|
lipocalin 6 |
| chr14_-_106803221 | 1.31 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr20_+_46008900 | 1.30 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 |
| chr1_-_1779976 | 1.29 |
ENST00000378625.5
|
NADK
|
NAD kinase |
| chr19_-_54364863 | 1.29 |
ENST00000348231.8
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1 |
| chr12_-_52517929 | 1.29 |
ENST00000548409.5
|
KRT5
|
keratin 5 |
| chr17_-_73227700 | 1.28 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104 member A |
| chr19_-_7702139 | 1.28 |
ENST00000346664.9
|
FCER2
|
Fc fragment of IgE receptor II |
| chr19_-_54364983 | 1.26 |
ENST00000434277.6
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1 |
| chrX_+_109536832 | 1.26 |
ENST00000372106.6
|
NXT2
|
nuclear transport factor 2 like export factor 2 |
| chr17_+_42851167 | 1.25 |
ENST00000613571.1
ENST00000308423.7 |
AOC3
|
amine oxidase copper containing 3 |
| chr3_-_108222383 | 1.25 |
ENST00000264538.4
|
IFT57
|
intraflagellar transport 57 |
| chr19_+_48019726 | 1.24 |
ENST00000593413.1
|
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr14_-_106117159 | 1.24 |
ENST00000390601.3
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 |
| chr5_+_150778781 | 1.24 |
ENST00000648745.1
|
SMIM3
|
small integral membrane protein 3 |
| chr4_-_48134200 | 1.24 |
ENST00000264316.9
|
TXK
|
TXK tyrosine kinase |
| chr12_+_10307818 | 1.23 |
ENST00000350274.9
ENST00000336164.9 |
KLRD1
|
killer cell lectin like receptor D1 |
| chr11_-_26572130 | 1.22 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr3_-_151203201 | 1.21 |
ENST00000480322.1
ENST00000309180.6 |
GPR171
|
G protein-coupled receptor 171 |
| chr1_-_108192818 | 1.20 |
ENST00000370041.4
|
SLC25A24
|
solute carrier family 25 member 24 |
| chr7_+_142670734 | 1.20 |
ENST00000390398.3
|
TRBV25-1
|
T cell receptor beta variable 25-1 |
| chr1_+_220879434 | 1.20 |
ENST00000366903.8
|
HLX
|
H2.0 like homeobox |
| chr14_+_75522427 | 1.19 |
ENST00000286639.8
|
BATF
|
basic leucine zipper ATF-like transcription factor |
| chr19_+_35448251 | 1.17 |
ENST00000599180.3
|
FFAR2
|
free fatty acid receptor 2 |
| chr14_-_106675544 | 1.16 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr5_+_74766981 | 1.15 |
ENST00000296802.9
|
NSA2
|
NSA2 ribosome biogenesis factor |
| chr6_-_41941507 | 1.15 |
ENST00000372987.8
|
CCND3
|
cyclin D3 |
| chr16_+_68085861 | 1.14 |
ENST00000570212.5
ENST00000562926.5 |
NFATC3
|
nuclear factor of activated T cells 3 |
| chr11_-_26572254 | 1.13 |
ENST00000529533.6
|
MUC15
|
mucin 15, cell surface associated |
| chr11_+_809961 | 1.13 |
ENST00000321153.9
ENST00000530797.5 |
RPLP2
|
ribosomal protein lateral stalk subunit P2 |
| chr19_-_54364908 | 1.12 |
ENST00000391742.7
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1 |
| chr1_+_156147350 | 1.12 |
ENST00000435124.5
ENST00000633494.1 |
SEMA4A
|
semaphorin 4A |
| chr17_+_60422483 | 1.11 |
ENST00000269127.5
|
C17orf64
|
chromosome 17 open reading frame 64 |
| chrX_+_72938163 | 1.11 |
ENST00000593662.1
ENST00000611003.2 |
FAM236A
|
family with sequence similarity 235 member A |
| chr14_-_106360320 | 1.11 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
| chr11_-_19202004 | 1.11 |
ENST00000648719.1
|
CSRP3
|
cysteine and glycine rich protein 3 |
| chr15_+_81296913 | 1.11 |
ENST00000394652.6
|
IL16
|
interleukin 16 |
| chr9_-_34710069 | 1.10 |
ENST00000378792.1
ENST00000259607.7 |
CCL21
|
C-C motif chemokine ligand 21 |
| chr16_+_85908988 | 1.10 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8 |
| chr2_+_202634960 | 1.10 |
ENST00000392238.3
|
FAM117B
|
family with sequence similarity 117 member B |
| chr19_-_14518383 | 1.09 |
ENST00000254322.3
ENST00000595139.2 |
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr5_-_140673568 | 1.09 |
ENST00000542735.2
|
DND1
|
DND microRNA-mediated repression inhibitor 1 |
| chr16_-_3256587 | 1.09 |
ENST00000536379.5
ENST00000541159.5 ENST00000339854.8 ENST00000219596.6 |
MEFV
|
MEFV innate immuity regulator, pyrin |
| chr19_-_344786 | 1.08 |
ENST00000264819.7
|
MIER2
|
MIER family member 2 |
| chr16_+_28985251 | 1.07 |
ENST00000360872.9
ENST00000566177.5 |
LAT
|
linker for activation of T cells |
| chr22_+_43151931 | 1.07 |
ENST00000329563.8
ENST00000396265.4 |
TSPO
|
translocator protein |
| chrX_-_42778155 | 1.07 |
ENST00000378131.4
|
PPP1R2C
|
PPP1R2C family member C |
| chr3_+_189100102 | 1.07 |
ENST00000412373.5
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr19_-_54364807 | 1.06 |
ENST00000474878.5
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1 |
| chr22_-_26565362 | 1.06 |
ENST00000398110.6
|
TPST2
|
tyrosylprotein sulfotransferase 2 |
| chr14_-_106088573 | 1.04 |
ENST00000632099.1
|
IGHV3-64D
|
immunoglobulin heavy variable 3-64D |
| chr16_+_21158653 | 1.04 |
ENST00000572258.5
ENST00000233047.9 ENST00000261388.7 ENST00000451578.6 ENST00000572599.5 ENST00000577162.1 |
TMEM159
|
transmembrane protein 159 |
| chr1_-_161307420 | 1.04 |
ENST00000491222.5
ENST00000672287.2 |
MPZ
|
myelin protein zero |
| chr6_-_131628165 | 1.03 |
ENST00000368053.8
ENST00000354577.8 ENST00000368060.7 ENST00000368068.8 |
MED23
|
mediator complex subunit 23 |
| chr15_-_55270874 | 1.03 |
ENST00000567380.5
ENST00000565972.5 ENST00000569493.5 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr22_+_36913620 | 1.03 |
ENST00000403662.8
ENST00000262825.9 |
CSF2RB
|
colony stimulating factor 2 receptor subunit beta |
| chr1_+_206897435 | 1.02 |
ENST00000391929.7
ENST00000294984.7 ENST00000611909.4 ENST00000367093.3 |
IL24
|
interleukin 24 |
| chr1_+_220786853 | 1.02 |
ENST00000366910.10
|
MTARC1
|
mitochondrial amidoxime reducing component 1 |
| chr9_+_70043840 | 1.02 |
ENST00000377182.5
|
MAMDC2
|
MAM domain containing 2 |
| chr6_-_32816910 | 1.02 |
ENST00000447394.1
ENST00000438763.7 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr1_+_26831992 | 1.01 |
ENST00000374141.6
|
ZDHHC18
|
zinc finger DHHC-type palmitoyltransferase 18 |
| chr2_+_231592858 | 1.00 |
ENST00000313965.4
|
TEX44
|
testis expressed 44 |
| chrX_-_72305892 | 1.00 |
ENST00000450875.5
ENST00000417400.1 ENST00000431381.5 ENST00000445983.5 ENST00000651998.1 |
CITED1
|
Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 1 |
| chr16_+_1966820 | 1.00 |
ENST00000569210.6
ENST00000569714.6 |
RNF151
|
ring finger protein 151 |
| chr5_-_41071314 | 0.99 |
ENST00000399564.5
|
MROH2B
|
maestro heat like repeat family member 2B |
| chr14_-_106593319 | 0.99 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr5_-_43043170 | 0.98 |
ENST00000314890.3
|
ANXA2R
|
annexin A2 receptor |
| chr17_+_47253817 | 0.97 |
ENST00000559488.7
ENST00000571680.1 |
ITGB3
|
integrin subunit beta 3 |
| chrX_-_48919015 | 0.95 |
ENST00000376509.4
|
PIM2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr19_-_42302766 | 0.95 |
ENST00000595530.5
ENST00000538771.5 ENST00000601865.5 |
PAFAH1B3
|
platelet activating factor acetylhydrolase 1b catalytic subunit 3 |
| chr1_-_182400660 | 0.95 |
ENST00000367565.2
|
TEDDM1
|
transmembrane epididymal protein 1 |
| chr19_+_47309853 | 0.95 |
ENST00000355085.4
|
C5AR1
|
complement C5a receptor 1 |
| chr16_+_28985043 | 0.95 |
ENST00000395456.7
ENST00000564277.5 ENST00000630764.2 ENST00000354453.7 |
LAT
|
linker for activation of T cells |
| chr21_+_41361999 | 0.95 |
ENST00000436410.5
ENST00000435611.6 ENST00000330714.8 |
MX2
|
MX dynamin like GTPase 2 |
| chrX_+_15749848 | 0.94 |
ENST00000479740.5
ENST00000454127.2 |
CA5B
|
carbonic anhydrase 5B |
| chr8_-_56446572 | 0.94 |
ENST00000518974.5
ENST00000451791.7 ENST00000523051.5 ENST00000518770.1 |
PENK
|
proenkephalin |
| chr12_+_6789508 | 0.94 |
ENST00000011653.9
ENST00000541982.5 ENST00000539492.1 |
CD4
|
CD4 molecule |
| chr1_-_46616804 | 0.92 |
ENST00000531769.6
ENST00000319928.8 |
MKNK1
MOB3C
|
MAPK interacting serine/threonine kinase 1 MOB kinase activator 3C |
| chrX_-_72782660 | 0.92 |
ENST00000596535.3
|
FAM236B
|
family with sequence similarity 236 member B |
| chr16_-_58546702 | 0.92 |
ENST00000567133.1
|
CNOT1
|
CCR4-NOT transcription complex subunit 1 |
| chr14_+_66486356 | 0.91 |
ENST00000636229.1
|
CCDC196
|
coiled-coil domain containing 196 |
| chr1_-_201421718 | 0.90 |
ENST00000367312.5
ENST00000555340.6 ENST00000361379.9 ENST00000622580.4 |
TNNI1
|
troponin I1, slow skeletal type |
| chr19_-_14517425 | 0.89 |
ENST00000676577.1
ENST00000677204.1 ENST00000598235.2 |
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr13_-_48444653 | 0.89 |
ENST00000378434.8
|
LPAR6
|
lysophosphatidic acid receptor 6 |
| chr8_-_56445941 | 0.88 |
ENST00000517415.1
ENST00000314922.3 |
PENK
|
proenkephalin |
| chr10_-_114684612 | 0.88 |
ENST00000533213.6
ENST00000369252.8 |
ABLIM1
|
actin binding LIM protein 1 |
| chr1_+_26832013 | 0.88 |
ENST00000534643.5
|
ZDHHC18
|
zinc finger DHHC-type palmitoyltransferase 18 |
| chr11_+_60378475 | 0.87 |
ENST00000358246.5
|
MS4A7
|
membrane spanning 4-domains A7 |
| chr17_-_56833889 | 0.87 |
ENST00000397861.7
|
C17orf67
|
chromosome 17 open reading frame 67 |
| chr15_+_34345865 | 0.86 |
ENST00000333756.4
|
NUTM1
|
NUT midline carcinoma family member 1 |
| chr18_-_21704763 | 0.85 |
ENST00000580981.5
ENST00000289119.7 |
ABHD3
|
abhydrolase domain containing 3, phospholipase |
| chr16_+_4556335 | 0.85 |
ENST00000444310.5
|
C16orf96
|
chromosome 16 open reading frame 96 |
| chr1_-_178871022 | 0.83 |
ENST00000367629.1
|
ANGPTL1
|
angiopoietin like 1 |
| chrX_-_155334580 | 0.83 |
ENST00000369449.7
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr19_-_17847962 | 0.82 |
ENST00000458235.7
ENST00000534444.1 |
JAK3
|
Janus kinase 3 |
| chr21_+_34073569 | 0.82 |
ENST00000399312.3
ENST00000381151.5 ENST00000362077.4 |
MRPS6
SLC5A3
ENSG00000272657.1
|
mitochondrial ribosomal protein S6 solute carrier family 5 member 3 novel transcript |
| chr10_+_116324440 | 0.82 |
ENST00000333254.4
|
CCDC172
|
coiled-coil domain containing 172 |
| chr12_+_99647749 | 0.81 |
ENST00000324341.2
|
FAM71C
|
family with sequence similarity 71 member C |
| chr9_-_19033200 | 0.81 |
ENST00000380534.9
ENST00000380530.1 |
SAXO1
|
stabilizer of axonemal microtubules 1 |
| chr22_+_35381086 | 0.80 |
ENST00000216117.9
ENST00000677931.1 ENST00000679074.1 |
HMOX1
|
heme oxygenase 1 |
| chr19_-_42217667 | 0.80 |
ENST00000336034.8
ENST00000596251.6 ENST00000598200.1 ENST00000598727.5 |
DEDD2
|
death effector domain containing 2 |
| chr16_+_28974813 | 0.80 |
ENST00000352260.11
|
SPNS1
|
sphingolipid transporter 1 (putative) |
| chr1_+_223393401 | 0.79 |
ENST00000366875.5
|
CCDC185
|
coiled-coil domain containing 185 |
| chr19_+_48552159 | 0.79 |
ENST00000201586.7
|
SULT2B1
|
sulfotransferase family 2B member 1 |
| chr7_-_117427487 | 0.78 |
ENST00000284629.7
|
ASZ1
|
ankyrin repeat, SAM and basic leucine zipper domain containing 1 |
| chr1_+_202214072 | 0.78 |
ENST00000439764.2
|
LGR6
|
leucine rich repeat containing G protein-coupled receptor 6 |
| chr14_-_24339951 | 0.78 |
ENST00000216274.10
|
RIPK3
|
receptor interacting serine/threonine kinase 3 |
| chr16_+_28974221 | 0.77 |
ENST00000567771.5
ENST00000568388.5 |
SPNS1
|
sphingolipid transporter 1 (putative) |
| chr7_-_112939773 | 0.77 |
ENST00000297145.9
|
BMT2
|
base methyltransferase of 25S rRNA 2 homolog |
| chr1_-_235866916 | 0.77 |
ENST00000389794.7
|
LYST
|
lysosomal trafficking regulator |
| chr1_-_178871060 | 0.76 |
ENST00000234816.7
|
ANGPTL1
|
angiopoietin like 1 |
| chr10_-_5499544 | 0.76 |
ENST00000380332.5
|
CALML5
|
calmodulin like 5 |
| chr2_+_189857393 | 0.76 |
ENST00000452382.1
|
PMS1
|
PMS1 homolog 1, mismatch repair system component |
| chr6_-_131628060 | 0.76 |
ENST00000539158.1
ENST00000368058.5 |
MED23
|
mediator complex subunit 23 |
| chr10_-_80356710 | 0.76 |
ENST00000454362.5
ENST00000372204.7 ENST00000372202.6 ENST00000453477.1 |
DYDC1
|
DPY30 domain containing 1 |
| chr2_+_74206384 | 0.75 |
ENST00000678623.1
ENST00000678731.1 |
MTHFD2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr4_-_76421121 | 0.75 |
ENST00000682701.1
|
CCDC158
|
coiled-coil domain containing 158 |
| chr1_-_235866867 | 0.74 |
ENST00000389793.7
|
LYST
|
lysosomal trafficking regulator |
| chr14_-_93976550 | 0.74 |
ENST00000555019.6
|
ASB2
|
ankyrin repeat and SOCS box containing 2 |
| chrX_+_109536641 | 0.74 |
ENST00000372107.5
|
NXT2
|
nuclear transport factor 2 like export factor 2 |
| chr2_-_197786238 | 0.73 |
ENST00000282278.12
|
BOLL
|
boule homolog, RNA binding protein |
| chr6_-_49964160 | 0.73 |
ENST00000322066.4
|
DEFB114
|
defensin beta 114 |
| chr1_-_156295647 | 0.73 |
ENST00000362007.6
ENST00000612353.4 ENST00000614643.4 ENST00000622703.4 |
GLMP
|
glycosylated lysosomal membrane protein |
| chr5_-_176537361 | 0.73 |
ENST00000274811.9
|
RNF44
|
ring finger protein 44 |
| chr11_+_59713403 | 0.73 |
ENST00000641815.1
|
STX3
|
syntaxin 3 |
| chr4_+_105895487 | 0.72 |
ENST00000506666.5
ENST00000503451.5 |
NPNT
|
nephronectin |
| chr6_+_152697888 | 0.72 |
ENST00000367245.5
ENST00000529453.1 |
MYCT1
|
MYC target 1 |
| chr19_-_474880 | 0.72 |
ENST00000382696.7
ENST00000315489.5 |
ODF3L2
|
outer dense fiber of sperm tails 3 like 2 |
| chr15_-_55289756 | 0.72 |
ENST00000336787.6
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr4_-_40630826 | 0.72 |
ENST00000505414.5
ENST00000511598.5 |
RBM47
|
RNA binding motif protein 47 |
| chr11_-_90223036 | 0.72 |
ENST00000320585.11
|
CHORDC1
|
cysteine and histidine rich domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 8.4 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.7 | 2.8 | GO:1900229 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.7 | 4.2 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.7 | 2.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.6 | 1.7 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.5 | 7.0 | GO:0072564 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.5 | 1.5 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.5 | 1.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.4 | 1.3 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.4 | 1.7 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.4 | 1.7 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.4 | 1.2 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.4 | 2.7 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.4 | 1.5 | GO:0052214 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.4 | 1.1 | GO:2000547 | mesangial cell-matrix adhesion(GO:0035759) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.4 | 1.8 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.3 | 1.4 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.3 | 1.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.3 | 0.6 | GO:0002339 | B cell selection(GO:0002339) |
| 0.3 | 0.9 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.3 | 0.9 | GO:0035397 | helper T cell enhancement of adaptive immune response(GO:0035397) |
| 0.3 | 1.9 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.3 | 1.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.3 | 2.9 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.3 | 0.8 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.3 | 1.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.3 | 0.8 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.3 | 1.8 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.3 | 0.8 | GO:2000452 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.2 | 1.7 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 1.7 | GO:0010266 | response to vitamin B1(GO:0010266) |
| 0.2 | 5.0 | GO:0044146 | negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.2 | 0.7 | GO:0002769 | natural killer cell inhibitory signaling pathway(GO:0002769) |
| 0.2 | 1.0 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.2 | 0.8 | GO:0009631 | cold acclimation(GO:0009631) |
| 0.2 | 1.0 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.2 | 0.8 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.2 | 0.4 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.2 | 0.9 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.2 | 1.6 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 0.7 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 0.5 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.2 | 1.7 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.2 | 0.7 | GO:0050705 | regulation of interleukin-1 alpha secretion(GO:0050705) |
| 0.2 | 0.8 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.9 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.7 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.7 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.1 | 1.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.7 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.9 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 0.7 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) renal protein absorption(GO:0097017) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.5 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 0.5 | GO:1904349 | regulation of small intestine smooth muscle contraction(GO:1904347) positive regulation of small intestine smooth muscle contraction(GO:1904349) small intestine smooth muscle contraction(GO:1990770) |
| 0.1 | 1.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.1 | 0.4 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.1 | 0.7 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 1.9 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.1 | 1.3 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 0.8 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 0.9 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.7 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.1 | 0.3 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.1 | 2.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.5 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 0.5 | GO:0090467 | L-arginine import(GO:0043091) divalent metal ion export(GO:0070839) arginine import(GO:0090467) |
| 0.1 | 0.7 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 1.0 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 1.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.2 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.1 | 1.4 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 1.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.4 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.1 | 0.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 2.0 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.3 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 0.2 | GO:0002537 | nitric oxide production involved in inflammatory response(GO:0002537) |
| 0.1 | 1.5 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 0.2 | GO:0002368 | B cell cytokine production(GO:0002368) |
| 0.1 | 1.7 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 1.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.1 | 2.4 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.1 | 0.8 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.3 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 0.6 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.3 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 0.4 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 1.6 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.9 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.1 | 0.3 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.8 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 0.9 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.6 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.1 | 0.2 | GO:0018963 | phthalate metabolic process(GO:0018963) epinephrine biosynthetic process(GO:0042418) phytoalexin metabolic process(GO:0052314) |
| 0.1 | 0.7 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.1 | 0.3 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 0.7 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.3 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.2 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.1 | 0.3 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.6 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.7 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.1 | 0.2 | GO:0036290 | protein trans-autophosphorylation(GO:0036290) peptidyl-serine trans-autophosphorylation(GO:1990579) |
| 0.1 | 0.5 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.1 | 0.8 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.3 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 0.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 1.0 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.3 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 0.4 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 1.2 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.1 | 1.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 4.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 2.5 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.9 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.2 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.1 | 2.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.4 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.1 | 1.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.8 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.1 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.6 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.1 | 1.2 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 1.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.2 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.1 | 2.8 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 1.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 1.1 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.7 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.5 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.3 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.6 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.4 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.8 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.7 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 1.1 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.7 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 1.8 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 1.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 4.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.3 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.9 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 0.4 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 1.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.3 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.4 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.5 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 1.0 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.0 | 0.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 4.0 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 1.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 1.2 | GO:0007620 | copulation(GO:0007620) |
| 0.0 | 0.4 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 1.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.7 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.5 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.0 | 1.4 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.8 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 1.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.3 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 | 1.4 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.6 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 6.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.9 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.7 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.0 | 0.8 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 1.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.9 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 2.2 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.7 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.0 | 0.3 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 1.7 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 2.3 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.1 | GO:0015881 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.0 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.4 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.1 | GO:0006463 | steroid hormone receptor complex assembly(GO:0006463) |
| 0.0 | 2.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.8 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.2 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.2 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 1.0 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.7 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.6 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 1.0 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 1.0 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 2.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.6 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.5 | GO:0045620 | negative regulation of lymphocyte differentiation(GO:0045620) |
| 0.0 | 1.6 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.0 | 0.3 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 0.2 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.0 | 0.4 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 1.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.8 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.5 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 2.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.5 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.7 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 1.1 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.7 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:1904948 | midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.0 | 0.4 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 1.0 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 4.3 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.4 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.3 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.2 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.5 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.3 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.4 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:1990834 | response to odorant(GO:1990834) |
| 0.0 | 2.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.1 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.4 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.1 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.5 | 3.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 0.3 | 1.0 | GO:0030526 | granulocyte macrophage colony-stimulating factor receptor complex(GO:0030526) |
| 0.3 | 1.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.3 | 1.5 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.3 | 1.8 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.2 | 2.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 0.6 | GO:0030849 | autosome(GO:0030849) |
| 0.2 | 2.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 1.4 | GO:0071546 | pi-body(GO:0071546) |
| 0.2 | 3.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 0.5 | GO:0097414 | glial cytoplasmic inclusion(GO:0097409) classical Lewy body(GO:0097414) Lewy neurite(GO:0097462) Lewy body corona(GO:1990038) |
| 0.1 | 0.7 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 7.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.9 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 1.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 1.6 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 6.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.7 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 1.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.8 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.7 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.3 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.1 | 4.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.8 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 2.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 1.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.3 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.1 | 0.2 | GO:1990332 | Ire1 complex(GO:1990332) |
| 0.1 | 1.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 2.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.5 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.3 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.1 | 0.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.5 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 3.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.6 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.3 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 9.8 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 7.4 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 2.6 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.9 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 2.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.8 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.5 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 4.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 8.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 2.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 1.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 2.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 2.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 2.0 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 1.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 3.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 1.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.2 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.6 | 3.0 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.6 | 7.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.4 | 1.3 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.4 | 4.4 | GO:0019863 | IgE binding(GO:0019863) |
| 0.4 | 2.0 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.4 | 4.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.3 | 0.9 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.3 | 1.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.3 | 0.8 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.3 | 1.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 1.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.3 | 1.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 0.8 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.2 | 1.7 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.2 | 1.0 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 1.7 | GO:0005497 | androgen binding(GO:0005497) |
| 0.2 | 0.7 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.2 | 0.5 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 1.6 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 2.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 1.5 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 2.9 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 2.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 0.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.7 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 2.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.7 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 1.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.7 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.4 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.1 | 0.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.4 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.5 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 5.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.4 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 1.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.5 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 0.5 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 2.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.1 | 1.3 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.1 | 0.8 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 0.8 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.5 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.9 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 1.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 0.7 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.1 | 0.4 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 1.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 1.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 1.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.3 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.1 | 0.6 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.2 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.1 | 1.7 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.8 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 4.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.6 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 0.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.1 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.1 | 1.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.5 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.4 | GO:0003916 | DNA topoisomerase activity(GO:0003916) DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.2 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 1.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.6 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.2 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.0 | 0.1 | GO:0030290 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.0 | 0.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 1.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 8.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.4 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 1.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.6 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.5 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 2.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.5 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 1.0 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 1.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.7 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.0 | 0.8 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 4.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 1.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 1.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 2.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 3.7 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.0 | 0.3 | GO:0001164 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.2 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 1.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.9 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.6 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.8 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 4.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 1.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.0 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 2.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 3.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 5.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 2.5 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 2.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 3.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 5.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 2.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.7 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 3.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 2.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 1.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 7.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 2.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 3.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 11.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 3.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 2.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 2.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 3.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.9 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 3.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 1.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.9 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 2.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.0 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 2.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 1.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.3 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 2.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |