Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
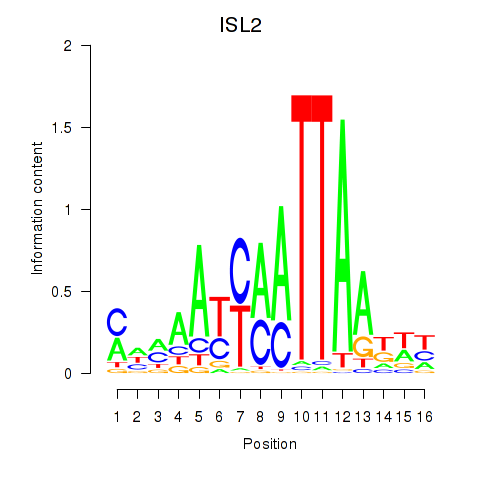
Results for ISL2
Z-value: 0.80
Transcription factors associated with ISL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ISL2
|
ENSG00000159556.10 | ISL LIM homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ISL2 | hg38_v1_chr15_+_76336755_76336779 | 0.15 | 4.1e-01 | Click! |
Activity profile of ISL2 motif
Sorted Z-values of ISL2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_55182442 | 1.91 |
ENST00000545075.3
|
MTRNR2L10
|
MT-RNR2 like 10 |
| chr5_+_40909490 | 1.76 |
ENST00000313164.10
|
C7
|
complement C7 |
| chr1_-_237945275 | 1.69 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2 like 11 |
| chr11_-_13495984 | 1.64 |
ENST00000282091.6
|
PTH
|
parathyroid hormone |
| chr10_-_29634964 | 1.64 |
ENST00000375398.6
ENST00000355867.8 |
SVIL
|
supervillin |
| chr11_-_13496018 | 1.34 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chrX_-_85379659 | 1.32 |
ENST00000262753.9
|
POF1B
|
POF1B actin binding protein |
| chr2_+_233760265 | 1.31 |
ENST00000305208.10
ENST00000360418.4 |
UGT1A1
|
UDP glucuronosyltransferase family 1 member A1 |
| chr2_-_189582012 | 1.31 |
ENST00000427419.5
ENST00000455320.5 |
SLC40A1
|
solute carrier family 40 member 1 |
| chr17_-_66229380 | 1.25 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr1_+_62597510 | 1.22 |
ENST00000371129.4
|
ANGPTL3
|
angiopoietin like 3 |
| chr21_+_38272410 | 1.20 |
ENST00000398934.5
ENST00000398930.5 |
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr1_-_178871022 | 1.13 |
ENST00000367629.1
|
ANGPTL1
|
angiopoietin like 1 |
| chr1_-_178871060 | 1.12 |
ENST00000234816.7
|
ANGPTL1
|
angiopoietin like 1 |
| chr8_+_91249307 | 1.11 |
ENST00000309536.6
ENST00000276609.8 |
SLC26A7
|
solute carrier family 26 member 7 |
| chr4_+_112647059 | 1.06 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein 7, transcriptional regulator |
| chr2_+_233671879 | 1.05 |
ENST00000354728.5
|
UGT1A9
|
UDP glucuronosyltransferase family 1 member A9 |
| chr7_+_123601815 | 1.05 |
ENST00000451215.6
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr2_-_162152239 | 1.05 |
ENST00000418842.7
|
GCG
|
glucagon |
| chr2_+_167135901 | 1.05 |
ENST00000628543.2
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr8_+_132897674 | 1.03 |
ENST00000518505.1
|
TG
|
thyroglobulin |
| chr7_+_123601836 | 1.03 |
ENST00000434204.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr2_-_162152404 | 1.01 |
ENST00000375497.3
|
GCG
|
glucagon |
| chr7_-_120857124 | 0.97 |
ENST00000441017.5
ENST00000424710.5 ENST00000433758.5 |
TSPAN12
|
tetraspanin 12 |
| chr15_+_58410543 | 0.95 |
ENST00000356113.10
ENST00000414170.7 |
LIPC
|
lipase C, hepatic type |
| chr14_-_67412112 | 0.94 |
ENST00000216446.9
|
PLEK2
|
pleckstrin 2 |
| chr10_-_93482326 | 0.92 |
ENST00000359263.9
|
MYOF
|
myoferlin |
| chr7_+_123601859 | 0.87 |
ENST00000437535.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr10_-_93482194 | 0.84 |
ENST00000358334.9
ENST00000371488.3 |
MYOF
|
myoferlin |
| chr19_+_14583076 | 0.83 |
ENST00000547437.5
ENST00000417570.6 |
CLEC17A
|
C-type lectin domain containing 17A |
| chrX_-_85379703 | 0.82 |
ENST00000373145.3
|
POF1B
|
POF1B actin binding protein |
| chr4_+_94207596 | 0.82 |
ENST00000359052.8
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr7_-_16804987 | 0.81 |
ENST00000401412.5
ENST00000419304.7 |
AGR2
|
anterior gradient 2, protein disulphide isomerase family member |
| chr10_-_93482287 | 0.80 |
ENST00000371489.5
|
MYOF
|
myoferlin |
| chr5_+_132873660 | 0.80 |
ENST00000296877.3
|
LEAP2
|
liver enriched antimicrobial peptide 2 |
| chr15_+_90191913 | 0.79 |
ENST00000559792.5
|
SEMA4B
|
semaphorin 4B |
| chr20_+_31514410 | 0.78 |
ENST00000335574.10
ENST00000340852.9 ENST00000398174.9 ENST00000466766.2 ENST00000498035.5 ENST00000344042.5 |
HM13
|
histocompatibility minor 13 |
| chrX_-_21658324 | 0.71 |
ENST00000379499.3
|
KLHL34
|
kelch like family member 34 |
| chr2_+_233917371 | 0.70 |
ENST00000324695.9
ENST00000433712.6 |
TRPM8
|
transient receptor potential cation channel subfamily M member 8 |
| chr6_+_130018565 | 0.68 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr2_-_10447771 | 0.68 |
ENST00000405333.5
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr15_+_64387828 | 0.66 |
ENST00000261884.8
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr5_+_40841308 | 0.63 |
ENST00000381677.4
ENST00000254691.10 |
CARD6
|
caspase recruitment domain family member 6 |
| chr12_+_130953898 | 0.61 |
ENST00000261654.10
|
ADGRD1
|
adhesion G protein-coupled receptor D1 |
| chr4_-_174263080 | 0.60 |
ENST00000615392.4
|
FBXO8
|
F-box protein 8 |
| chr16_+_28553908 | 0.59 |
ENST00000317058.8
|
SGF29
|
SAGA complex associated factor 29 |
| chr3_-_122022122 | 0.59 |
ENST00000393631.5
ENST00000273691.7 ENST00000344209.10 |
ILDR1
|
immunoglobulin like domain containing receptor 1 |
| chrX_-_31266857 | 0.58 |
ENST00000378702.8
ENST00000361471.8 |
DMD
|
dystrophin |
| chr11_-_8871385 | 0.56 |
ENST00000527516.5
ENST00000533471.5 |
DENND2B
|
DENN domain containing 2B |
| chr4_+_85604146 | 0.56 |
ENST00000512201.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr3_+_136930469 | 0.56 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr2_+_87748087 | 0.56 |
ENST00000359481.9
|
PLGLB2
|
plasminogen like B2 |
| chr2_-_157439403 | 0.55 |
ENST00000418920.5
|
CYTIP
|
cytohesin 1 interacting protein |
| chrX_-_15314543 | 0.55 |
ENST00000344384.8
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chrX_-_31266925 | 0.54 |
ENST00000680557.1
ENST00000378680.6 ENST00000378723.7 ENST00000680768.2 ENST00000681870.1 ENST00000680355.1 ENST00000682322.1 ENST00000682600.1 ENST00000680162.2 ENST00000681153.1 ENST00000681334.1 |
DMD
|
dystrophin |
| chr14_+_21997531 | 0.54 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr7_-_120858066 | 0.51 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12 |
| chr13_+_48976597 | 0.51 |
ENST00000541916.5
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr20_-_35147285 | 0.50 |
ENST00000374491.3
ENST00000374492.8 |
EDEM2
|
ER degradation enhancing alpha-mannosidase like protein 2 |
| chr2_+_86441995 | 0.49 |
ENST00000427678.1
|
KDM3A
|
lysine demethylase 3A |
| chrX_-_32155462 | 0.49 |
ENST00000359836.5
ENST00000378707.7 ENST00000541735.5 ENST00000684130.1 ENST00000682238.1 ENST00000620040.5 ENST00000474231.5 |
DMD
|
dystrophin |
| chr21_-_30492008 | 0.46 |
ENST00000334063.6
|
KRTAP19-3
|
keratin associated protein 19-3 |
| chr4_-_48114523 | 0.46 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr3_-_142029108 | 0.45 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr11_-_28108109 | 0.45 |
ENST00000263181.7
|
KIF18A
|
kinesin family member 18A |
| chr4_+_168497044 | 0.44 |
ENST00000505667.6
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr7_+_16661182 | 0.44 |
ENST00000446596.5
ENST00000452975.6 ENST00000438834.5 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr19_-_45730861 | 0.44 |
ENST00000317683.4
|
FBXO46
|
F-box protein 46 |
| chr6_+_54099538 | 0.43 |
ENST00000447836.6
|
MLIP
|
muscular LMNA interacting protein |
| chr7_-_116030750 | 0.43 |
ENST00000265440.12
ENST00000320239.11 |
TFEC
|
transcription factor EC |
| chr2_-_40453438 | 0.42 |
ENST00000455476.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr19_+_3185911 | 0.42 |
ENST00000246117.9
ENST00000588428.5 |
NCLN
|
nicalin |
| chr14_+_22202561 | 0.42 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr7_-_116030735 | 0.42 |
ENST00000393485.5
|
TFEC
|
transcription factor EC |
| chr12_-_56741535 | 0.42 |
ENST00000647707.1
|
ENSG00000285625.1
|
novel protein |
| chr1_-_225428813 | 0.41 |
ENST00000338179.6
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr12_-_57818704 | 0.41 |
ENST00000549994.1
|
AVIL
|
advillin |
| chr2_-_169031317 | 0.41 |
ENST00000650372.1
|
ABCB11
|
ATP binding cassette subfamily B member 11 |
| chr4_+_168497066 | 0.41 |
ENST00000261509.10
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr17_-_49709014 | 0.40 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35 member B1 |
| chr11_+_28108248 | 0.40 |
ENST00000406787.7
ENST00000403099.5 ENST00000407364.8 |
METTL15
|
methyltransferase like 15 |
| chr3_+_186974957 | 0.40 |
ENST00000438590.5
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr4_-_67963441 | 0.39 |
ENST00000508048.6
|
TMPRSS11A
|
transmembrane serine protease 11A |
| chr12_+_64405046 | 0.37 |
ENST00000540203.5
|
XPOT
|
exportin for tRNA |
| chr6_+_31137646 | 0.37 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr19_+_49513353 | 0.36 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr4_+_25160631 | 0.36 |
ENST00000510415.1
ENST00000507794.2 ENST00000512921.4 |
SEPSECS-AS1
PI4K2B
|
SEPSECS antisense RNA 1 (head to head) phosphatidylinositol 4-kinase type 2 beta |
| chrX_+_83861126 | 0.36 |
ENST00000621735.4
ENST00000329312.5 |
CYLC1
|
cylicin 1 |
| chr8_-_123729211 | 0.36 |
ENST00000520519.1
|
ANXA13
|
annexin A13 |
| chr4_+_94207845 | 0.36 |
ENST00000457823.6
ENST00000354268.9 |
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr20_-_52105644 | 0.35 |
ENST00000371523.8
|
ZFP64
|
ZFP64 zinc finger protein |
| chr7_-_45111673 | 0.34 |
ENST00000461363.1
ENST00000258770.8 ENST00000495078.1 ENST00000494076.5 ENST00000478532.5 ENST00000361278.7 |
TBRG4
|
transforming growth factor beta regulator 4 |
| chr19_+_48695952 | 0.34 |
ENST00000522966.2
ENST00000425340.3 ENST00000391876.5 |
FUT2
|
fucosyltransferase 2 |
| chr9_+_72577369 | 0.33 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr8_+_69492793 | 0.33 |
ENST00000616868.1
ENST00000419716.7 ENST00000402687.9 |
SULF1
|
sulfatase 1 |
| chr5_+_102865244 | 0.33 |
ENST00000511477.5
ENST00000506006.1 ENST00000509832.5 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr20_+_45420585 | 0.32 |
ENST00000639239.1
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis class T |
| chr2_-_87021844 | 0.32 |
ENST00000355705.4
ENST00000409310.6 |
PLGLB1
|
plasminogen like B1 |
| chr6_+_54099565 | 0.31 |
ENST00000511678.5
|
MLIP
|
muscular LMNA interacting protein |
| chr12_-_110499546 | 0.30 |
ENST00000552130.6
|
VPS29
|
VPS29 retromer complex component |
| chr10_-_114144599 | 0.30 |
ENST00000428953.1
|
CCDC186
|
coiled-coil domain containing 186 |
| chr6_+_63635792 | 0.29 |
ENST00000262043.8
ENST00000506783.5 ENST00000481385.6 ENST00000515594.5 ENST00000494284.6 |
PHF3
|
PHD finger protein 3 |
| chr11_-_26723361 | 0.29 |
ENST00000533617.5
|
SLC5A12
|
solute carrier family 5 member 12 |
| chr16_+_86566821 | 0.29 |
ENST00000649859.1
|
FOXC2
|
forkhead box C2 |
| chr2_-_84881238 | 0.29 |
ENST00000335459.9
|
TRABD2A
|
TraB domain containing 2A |
| chr14_+_34993240 | 0.29 |
ENST00000677647.1
|
SRP54
|
signal recognition particle 54 |
| chr5_+_58491451 | 0.28 |
ENST00000513924.2
ENST00000515443.2 |
GAPT
|
GRB2 binding adaptor protein, transmembrane |
| chr12_+_4720395 | 0.28 |
ENST00000252318.7
|
GALNT8
|
polypeptide N-acetylgalactosaminyltransferase 8 |
| chr4_-_103019634 | 0.28 |
ENST00000510559.1
ENST00000296422.12 ENST00000394789.7 |
SLC9B1
|
solute carrier family 9 member B1 |
| chr6_-_49964160 | 0.28 |
ENST00000322066.4
|
DEFB114
|
defensin beta 114 |
| chr5_+_58491427 | 0.28 |
ENST00000396776.6
ENST00000502276.6 ENST00000511930.2 |
GAPT
|
GRB2 binding adaptor protein, transmembrane |
| chr1_-_17439657 | 0.27 |
ENST00000375436.9
|
RCC2
|
regulator of chromosome condensation 2 |
| chr7_-_33100886 | 0.27 |
ENST00000448915.1
|
RP9
|
RP9 pre-mRNA splicing factor |
| chr13_-_46142834 | 0.27 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr1_-_169703329 | 0.27 |
ENST00000497295.1
|
SELL
|
selectin L |
| chr22_-_28711931 | 0.27 |
ENST00000434810.5
ENST00000456369.5 |
CHEK2
|
checkpoint kinase 2 |
| chrM_+_9207 | 0.26 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr6_-_111483700 | 0.26 |
ENST00000435970.5
ENST00000358835.7 |
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr4_-_137532452 | 0.26 |
ENST00000412923.6
ENST00000511115.5 ENST00000344876.9 ENST00000507846.5 ENST00000510305.5 ENST00000611581.1 |
PCDH18
|
protocadherin 18 |
| chr12_-_95551417 | 0.26 |
ENST00000258499.8
|
USP44
|
ubiquitin specific peptidase 44 |
| chr14_+_60982358 | 0.26 |
ENST00000526105.5
|
SLC38A6
|
solute carrier family 38 member 6 |
| chr17_-_10469558 | 0.25 |
ENST00000255381.2
|
MYH4
|
myosin heavy chain 4 |
| chr2_+_127419686 | 0.25 |
ENST00000427769.5
|
PROC
|
protein C, inactivator of coagulation factors Va and VIIIa |
| chr1_-_247760556 | 0.25 |
ENST00000641256.1
|
OR1C1
|
olfactory receptor family 1 subfamily C member 1 |
| chr15_+_78077798 | 0.25 |
ENST00000409568.6
|
SH2D7
|
SH2 domain containing 7 |
| chr7_-_42152396 | 0.24 |
ENST00000642432.1
ENST00000647255.1 ENST00000677288.1 |
GLI3
|
GLI family zinc finger 3 |
| chr6_-_155455830 | 0.24 |
ENST00000159060.3
|
NOX3
|
NADPH oxidase 3 |
| chr3_-_150703965 | 0.23 |
ENST00000498386.1
|
ERICH6
|
glutamate rich 6 |
| chr4_+_155854758 | 0.22 |
ENST00000506072.5
ENST00000507590.5 |
TDO2
|
tryptophan 2,3-dioxygenase |
| chr17_+_10697576 | 0.22 |
ENST00000379774.5
|
ADPRM
|
ADP-ribose/CDP-alcohol diphosphatase, manganese dependent |
| chr17_-_7404039 | 0.21 |
ENST00000576017.1
ENST00000302422.4 |
TMEM256
|
transmembrane protein 256 |
| chr3_-_186544377 | 0.21 |
ENST00000307944.6
|
CRYGS
|
crystallin gamma S |
| chr11_+_55723776 | 0.21 |
ENST00000641440.1
|
OR5D3P
|
olfactory receptor family 5 subfamily D member 3 pseudogene |
| chr17_-_40999903 | 0.20 |
ENST00000391587.3
|
KRTAP3-2
|
keratin associated protein 3-2 |
| chr11_+_5389377 | 0.20 |
ENST00000328611.5
|
OR51M1
|
olfactory receptor family 51 subfamily M member 1 |
| chr12_-_118190510 | 0.20 |
ENST00000540561.5
ENST00000537952.1 ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr14_+_22096017 | 0.20 |
ENST00000390452.2
|
TRDV1
|
T cell receptor delta variable 1 |
| chrM_+_10055 | 0.19 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 3 |
| chr7_-_111392915 | 0.19 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr17_+_7252502 | 0.19 |
ENST00000570322.5
ENST00000576496.5 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chrY_-_23694579 | 0.19 |
ENST00000343584.10
|
PRYP3
|
PTPN13 like Y-linked pseudogene 3 |
| chr17_+_1771688 | 0.18 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin family F member 1 |
| chr2_-_206213430 | 0.18 |
ENST00000447845.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr1_-_152806628 | 0.18 |
ENST00000606576.1
ENST00000607093.2 |
LCE1C
|
late cornified envelope 1C |
| chr17_-_8210203 | 0.18 |
ENST00000578549.5
ENST00000582368.5 |
AURKB
|
aurora kinase B |
| chr9_+_108862255 | 0.18 |
ENST00000333999.5
|
ACTL7A
|
actin like 7A |
| chr2_+_108239965 | 0.18 |
ENST00000681802.2
ENST00000329106.3 |
SULT1C3
|
sulfotransferase family 1C member 3 |
| chr3_-_191282383 | 0.17 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr4_-_119322128 | 0.17 |
ENST00000274024.4
|
FABP2
|
fatty acid binding protein 2 |
| chr17_-_64390592 | 0.17 |
ENST00000563523.5
|
PECAM1
|
platelet and endothelial cell adhesion molecule 1 |
| chr20_-_31390483 | 0.16 |
ENST00000376315.2
|
DEFB119
|
defensin beta 119 |
| chrY_+_18546691 | 0.16 |
ENST00000309834.8
ENST00000307393.3 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor Y-linked 1 |
| chr4_+_168497113 | 0.16 |
ENST00000511948.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr19_-_44529687 | 0.16 |
ENST00000621342.4
ENST00000617951.1 |
CEACAM20
|
CEA cell adhesion molecule 20 |
| chr11_-_5548540 | 0.15 |
ENST00000322653.7
|
OR52H1
|
olfactory receptor family 52 subfamily H member 1 |
| chr13_+_30422487 | 0.15 |
ENST00000638137.1
ENST00000635918.2 |
UBE2L5
|
ubiquitin conjugating enzyme E2 L5 |
| chr10_+_94762673 | 0.15 |
ENST00000480405.2
ENST00000371321.9 |
CYP2C19
|
cytochrome P450 family 2 subfamily C member 19 |
| chr14_+_21621801 | 0.15 |
ENST00000542354.1
|
TRAV1-1
|
T cell receptor alpha variable 1-1 |
| chr4_+_70242583 | 0.15 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr15_-_75455767 | 0.15 |
ENST00000360439.8
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr3_+_169911566 | 0.14 |
ENST00000428432.6
ENST00000335556.7 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr7_-_84492718 | 0.14 |
ENST00000424555.5
|
SEMA3A
|
semaphorin 3A |
| chr16_+_53099100 | 0.14 |
ENST00000565832.5
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr16_-_1488764 | 0.13 |
ENST00000447419.7
ENST00000440447.2 |
PTX4
|
pentraxin 4 |
| chr5_-_131165231 | 0.13 |
ENST00000675100.1
ENST00000304043.10 ENST00000513012.2 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr11_-_124320197 | 0.13 |
ENST00000624618.2
|
OR8D2
|
olfactory receptor family 8 subfamily D member 2 |
| chr17_+_7428831 | 0.12 |
ENST00000636696.4
ENST00000651079.1 |
SPEM3
|
SPEM family member 3 |
| chr14_+_19719015 | 0.12 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor family 4 subfamily N member 2 |
| chr14_-_55440616 | 0.12 |
ENST00000247219.5
|
TBPL2
|
TATA-box binding protein like 2 |
| chr6_-_138512493 | 0.12 |
ENST00000533765.1
|
NHSL1
|
NHS like 1 |
| chr11_+_35186820 | 0.11 |
ENST00000531110.6
ENST00000525685.6 |
CD44
|
CD44 molecule (Indian blood group) |
| chr11_-_16397521 | 0.10 |
ENST00000533411.5
|
SOX6
|
SRY-box transcription factor 6 |
| chr3_+_40477107 | 0.10 |
ENST00000314686.9
ENST00000447116.6 ENST00000429348.6 ENST00000432264.4 ENST00000456778.5 |
ZNF619
|
zinc finger protein 619 |
| chr12_+_55444069 | 0.10 |
ENST00000641516.1
ENST00000641202.1 |
OR6C2
|
olfactory receptor family 6 subfamily C member 2 |
| chr1_+_98661666 | 0.09 |
ENST00000529992.5
|
SNX7
|
sorting nexin 7 |
| chr12_+_1970772 | 0.09 |
ENST00000682544.1
|
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C |
| chr12_+_69825273 | 0.09 |
ENST00000547771.6
|
MYRFL
|
myelin regulatory factor like |
| chr17_-_7315312 | 0.08 |
ENST00000577040.2
ENST00000389167.9 ENST00000380728.7 |
GPS2
|
G protein pathway suppressor 2 |
| chr1_+_112718880 | 0.08 |
ENST00000361886.4
|
TAFA3
|
TAFA chemokine like family member 3 |
| chrX_-_101617921 | 0.08 |
ENST00000361910.9
ENST00000538627.5 ENST00000539247.5 |
ARMCX6
|
armadillo repeat containing X-linked 6 |
| chr1_+_13303539 | 0.08 |
ENST00000437300.2
|
PRAMEF33
|
PRAME family member 33 |
| chr1_+_149899561 | 0.08 |
ENST00000369152.6
|
BOLA1
|
bolA family member 1 |
| chr10_-_5003850 | 0.08 |
ENST00000421196.7
ENST00000455190.2 ENST00000380753.8 |
AKR1C2
|
aldo-keto reductase family 1 member C2 |
| chrX_-_72808210 | 0.07 |
ENST00000419795.6
ENST00000458170.1 |
FAM236D
|
family with sequence similarity 236 member D |
| chr2_-_206213362 | 0.07 |
ENST00000442134.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr15_+_48191648 | 0.07 |
ENST00000646012.1
ENST00000561127.5 ENST00000647546.1 ENST00000559641.5 ENST00000417307.3 |
SLC12A1
CTXN2
|
solute carrier family 12 member 1 cortexin 2 |
| chr5_+_111092172 | 0.07 |
ENST00000612402.4
|
WDR36
|
WD repeat domain 36 |
| chr13_+_23570370 | 0.07 |
ENST00000403372.6
ENST00000248484.9 |
TNFRSF19
|
TNF receptor superfamily member 19 |
| chr11_-_56614874 | 0.07 |
ENST00000641076.1
|
OR5M1
|
olfactory receptor family 5 subfamily M member 1 |
| chr2_-_158380960 | 0.07 |
ENST00000409187.5
|
CCDC148
|
coiled-coil domain containing 148 |
| chr1_-_13056575 | 0.06 |
ENST00000436041.6
|
PRAMEF27
|
PRAME family member 27 |
| chr11_-_62754141 | 0.06 |
ENST00000527994.1
ENST00000394807.5 ENST00000673933.1 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr11_-_75089754 | 0.06 |
ENST00000305159.3
|
OR2AT4
|
olfactory receptor family 2 subfamily AT member 4 |
| chr12_+_55492378 | 0.06 |
ENST00000548615.1
|
OR6C68
|
olfactory receptor family 6 subfamily C member 68 |
| chr11_-_63608542 | 0.06 |
ENST00000540943.1
|
PLAAT3
|
phospholipase A and acyltransferase 3 |
| chr1_+_149899618 | 0.06 |
ENST00000369150.1
|
BOLA1
|
bolA family member 1 |
| chr12_-_54295748 | 0.06 |
ENST00000540264.2
ENST00000312156.8 |
NFE2
|
nuclear factor, erythroid 2 |
| chr1_-_223665723 | 0.06 |
ENST00000366873.6
|
CAPN8
|
calpain 8 |
| chr12_+_1970809 | 0.06 |
ENST00000683781.1
ENST00000682462.1 |
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C |
| chr6_-_24358036 | 0.05 |
ENST00000378454.8
|
DCDC2
|
doublecortin domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ISL2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.4 | 1.3 | GO:1903988 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.3 | 2.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.3 | 1.3 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.3 | 0.8 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.2 | 0.6 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.2 | 0.5 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.2 | 0.8 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.1 | 2.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 1.1 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 0.5 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 0.4 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.7 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 1.2 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 1.2 | GO:0051005 | negative regulation of phospholipase activity(GO:0010519) negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.4 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 1.6 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.6 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.7 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.3 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.6 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 1.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.0 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.9 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.5 | GO:0072011 | glomerular endothelium development(GO:0072011) |
| 0.1 | 0.3 | GO:1904806 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.1 | 0.3 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 1.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.4 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.5 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.2 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.2 | GO:0060875 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.1 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.3 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.3 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.3 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 1.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 1.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.4 | GO:0071313 | cellular response to caffeine(GO:0071313) positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.6 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.2 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 1.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 2.1 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.2 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.3 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 1.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.3 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.8 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 0.4 | GO:0033150 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.1 | 1.6 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 1.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.3 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 1.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.9 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 2.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 1.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.2 | 1.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 0.6 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.4 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.4 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.8 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 3.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 2.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 2.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.3 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 3.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.0 | 0.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.3 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 4.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 2.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 1.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 3.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |