Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for JUN
Z-value: 2.03
Transcription factors associated with JUN
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JUN
|
ENSG00000177606.8 | Jun proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JUN | hg38_v1_chr1_-_58784035_58784054 | 0.02 | 9.2e-01 | Click! |
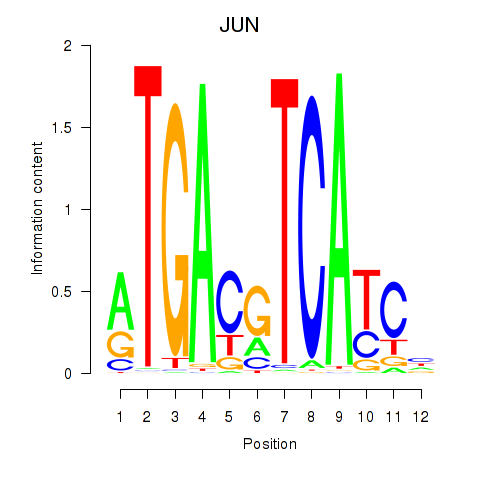
Activity profile of JUN motif
Sorted Z-values of JUN motif
Network of associatons between targets according to the STRING database.
First level regulatory network of JUN
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 9.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 1.5 | 25.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 1.4 | 9.9 | GO:0007343 | egg activation(GO:0007343) |
| 1.1 | 8.5 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 1.0 | 3.1 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 1.0 | 4.0 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.9 | 4.7 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.9 | 5.6 | GO:0032826 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.9 | 2.7 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.7 | 3.7 | GO:0006288 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) base-excision repair, DNA ligation(GO:0006288) |
| 0.7 | 2.8 | GO:0009631 | cold acclimation(GO:0009631) |
| 0.7 | 2.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.7 | 2.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.6 | 3.8 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.6 | 2.4 | GO:0072560 | type B pancreatic cell maturation(GO:0072560) |
| 0.6 | 3.4 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.6 | 3.4 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.5 | 3.7 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.5 | 13.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.5 | 2.3 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.5 | 2.7 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.4 | 1.8 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.4 | 2.2 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.4 | 3.4 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.4 | 1.3 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.4 | 3.0 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.4 | 3.7 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.4 | 0.4 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.4 | 3.9 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.4 | 1.5 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.4 | 1.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.4 | 1.4 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.4 | 1.8 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.4 | 4.6 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.3 | 1.7 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.3 | 1.0 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.3 | 1.4 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.3 | 1.0 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.3 | 0.9 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.3 | 0.6 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.3 | 1.5 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.3 | 1.2 | GO:0061163 | endoplasmic reticulum polarization(GO:0061163) actin filament bundle retrograde transport(GO:0061573) actin filament bundle distribution(GO:0070650) |
| 0.3 | 13.9 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.3 | 1.7 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.3 | 1.7 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.3 | 0.8 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.3 | 5.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.3 | 2.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) positive regulation of long term synaptic depression(GO:1900454) |
| 0.3 | 1.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.3 | 1.0 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.3 | 0.8 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.2 | 0.7 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.2 | 7.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 1.4 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 3.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 2.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.2 | 2.7 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.2 | 4.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 1.5 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 2.6 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 | 1.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 0.6 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.2 | 1.9 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.2 | 1.6 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.2 | 1.5 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.2 | 0.5 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.2 | 0.5 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.2 | 2.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 3.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 1.4 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.2 | 12.1 | GO:0030317 | sperm motility(GO:0030317) |
| 0.2 | 0.9 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 3.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 20.5 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 0.9 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 4.1 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.7 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 3.4 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 25.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.5 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 5.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.5 | GO:1900222 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.1 | 1.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 2.9 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.6 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.1 | 0.4 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.1 | 2.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.6 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 1.3 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.1 | 1.5 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 1.0 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.5 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.2 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.0 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.5 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 4.0 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 1.2 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.7 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 | 0.7 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 1.9 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.1 | 0.8 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.1 | 0.7 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.1 | 1.0 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.8 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 0.3 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.1 | 5.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.9 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.3 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 1.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.6 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 | 0.5 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 0.5 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 1.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 3.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 36.4 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 0.3 | GO:1904021 | negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.1 | 1.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 2.2 | GO:0000732 | strand displacement(GO:0000732) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 1.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.9 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 5.7 | GO:0043525 | positive regulation of neuron apoptotic process(GO:0043525) |
| 0.1 | 5.0 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 1.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.3 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 3.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.2 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.4 | GO:0045136 | development of secondary sexual characteristics(GO:0045136) development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.5 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 2.2 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.7 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.6 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 3.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.5 | GO:0007135 | meiosis II(GO:0007135) |
| 0.0 | 3.5 | GO:0010955 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 0.7 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 1.3 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 1.2 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 4.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 5.4 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 1.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 1.0 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.7 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:1904124 | negative regulation of norepinephrine secretion(GO:0010700) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 1.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.7 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.5 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 1.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.3 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.5 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 3.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.8 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 1.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 1.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.8 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.7 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 8.9 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.9 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 1.5 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.3 | GO:2000111 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) positive regulation of myeloid cell apoptotic process(GO:0033034) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.4 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.4 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:1901552 | negative regulation of excitatory postsynaptic potential(GO:0090394) positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.8 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.8 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 2.0 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 1.3 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.0 | 0.8 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.1 | GO:1904207 | chemokine (C-C motif) ligand 2 secretion(GO:0035926) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 15.8 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 1.9 | 3.8 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 1.8 | 10.9 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 1.0 | 6.7 | GO:0031523 | Myb complex(GO:0031523) |
| 0.9 | 13.9 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.8 | 9.8 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.8 | 4.6 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.5 | 4.8 | GO:0071546 | pi-body(GO:0071546) |
| 0.5 | 3.7 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.5 | 8.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.4 | 1.7 | GO:0097545 | axonemal outer doublet(GO:0097545) |
| 0.4 | 5.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 5.6 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.4 | 1.3 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 2.7 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.3 | 3.0 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 2.9 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 1.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.3 | 11.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.3 | 2.8 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.3 | 2.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 1.4 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 2.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.2 | 3.0 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 0.8 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 26.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 3.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 3.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.2 | 1.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 0.7 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.2 | 4.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 5.8 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.2 | 3.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 2.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 3.0 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 3.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 2.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.5 | GO:0071757 | hexameric IgM immunoglobulin complex(GO:0071757) |
| 0.1 | 0.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 2.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 19.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 1.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.9 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 2.8 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 3.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 6.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 1.0 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 1.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 2.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 5.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.9 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.0 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 7.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 0.9 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 2.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 1.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.6 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 4.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.9 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.7 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 2.1 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.8 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 2.0 | GO:0031902 | late endosome membrane(GO:0031902) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 25.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.9 | 3.8 | GO:0004040 | amidase activity(GO:0004040) |
| 0.9 | 4.5 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.7 | 5.8 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.7 | 3.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.7 | 2.0 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.5 | 3.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.5 | 3.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 6.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 4.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.4 | 3.9 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.3 | 1.0 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.3 | 3.5 | GO:0089720 | caspase binding(GO:0089720) |
| 0.3 | 1.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.3 | 2.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 4.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 1.4 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.3 | 4.8 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 3.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.3 | 1.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.3 | 1.3 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.2 | 2.0 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.2 | 2.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 2.7 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.2 | 2.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 0.6 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.2 | 1.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 0.6 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.2 | 1.8 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.2 | 1.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 0.7 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.2 | 0.7 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.2 | 0.5 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.2 | 5.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 1.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 3.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 4.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 2.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 2.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.4 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.1 | 3.0 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.1 | 1.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 1.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 5.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.0 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 2.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 3.7 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 1.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 2.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.7 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 4.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.5 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.9 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 1.0 | GO:0015180 | hydrogen:amino acid symporter activity(GO:0005280) L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.4 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.8 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 2.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.0 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 1.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 1.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 2.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 7.1 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 3.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 21.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.3 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.1 | 0.4 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 2.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 3.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.3 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 4.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 4.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 2.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 2.7 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.1 | 2.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 6.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.2 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 22.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.1 | 1.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 3.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 1.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 2.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.9 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 2.5 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 1.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 7.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 1.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 12.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 2.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0004605 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 2.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 1.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 1.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.5 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.9 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 1.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 12.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.7 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 6.1 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 1.3 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 4.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 3.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 5.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 3.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 2.8 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 5.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 2.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 6.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 3.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 2.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 2.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 2.6 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 3.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.7 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 2.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 8.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.2 | 2.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 3.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.2 | 7.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 3.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 3.8 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 2.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 4.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 4.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 3.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 3.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 2.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 2.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 2.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 2.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 2.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.9 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.6 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |