Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
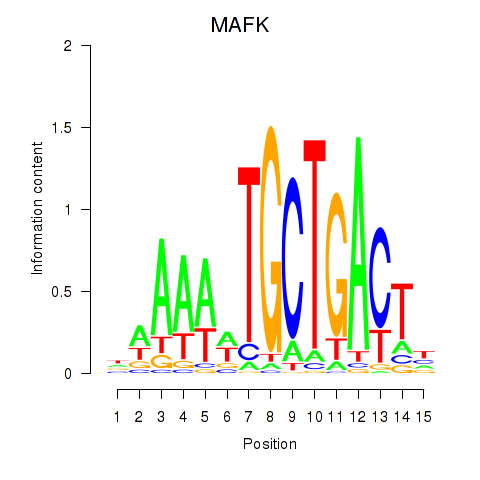
Results for MAFK
Z-value: 1.68
Transcription factors associated with MAFK
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFK
|
ENSG00000198517.10 | MAF bZIP transcription factor K |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MAFK | hg38_v1_chr7_+_1530684_1530726 | -0.21 | 2.4e-01 | Click! |
Activity profile of MAFK motif
Sorted Z-values of MAFK motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_88885397 | 12.34 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr1_+_198638457 | 8.99 |
ENST00000367379.6
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr14_+_20955484 | 8.09 |
ENST00000304625.3
|
RNASE2
|
ribonuclease A family member 2 |
| chr5_+_55102635 | 8.00 |
ENST00000274306.7
|
GZMA
|
granzyme A |
| chr1_+_198638723 | 7.85 |
ENST00000643513.1
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr1_+_198638968 | 7.57 |
ENST00000348564.11
ENST00000530727.5 ENST00000442510.8 ENST00000645247.1 ENST00000367367.8 ENST00000367364.5 ENST00000413409.6 |
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr1_+_158831323 | 7.19 |
ENST00000368141.5
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr14_-_106538331 | 6.82 |
ENST00000390624.3
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr7_+_142300924 | 5.72 |
ENST00000455382.2
|
TRBV2
|
T cell receptor beta variable 2 |
| chr1_-_169711603 | 5.61 |
ENST00000236147.6
ENST00000650983.1 |
SELL
|
selectin L |
| chr14_-_106235582 | 5.44 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr2_+_89913982 | 5.41 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr14_-_106557465 | 5.09 |
ENST00000390625.3
|
IGHV3-49
|
immunoglobulin heavy variable 3-49 |
| chr14_-_106038355 | 5.06 |
ENST00000390597.3
|
IGHV2-5
|
immunoglobulin heavy variable 2-5 |
| chr4_-_152679984 | 4.93 |
ENST00000304385.8
ENST00000504064.1 |
TMEM154
|
transmembrane protein 154 |
| chr19_-_54281082 | 4.83 |
ENST00000314446.10
|
LILRB2
|
leukocyte immunoglobulin like receptor B2 |
| chr2_-_89027700 | 4.66 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11 |
| chr3_-_39280432 | 4.63 |
ENST00000542107.5
ENST00000435290.1 |
CX3CR1
|
C-X3-C motif chemokine receptor 1 |
| chrX_+_124346544 | 4.62 |
ENST00000371139.9
|
SH2D1A
|
SH2 domain containing 1A |
| chr7_+_142320677 | 4.54 |
ENST00000390381.3
|
TRBV5-1
|
T cell receptor beta variable 5-1 |
| chr6_+_6588082 | 4.54 |
ENST00000379953.6
|
LY86
|
lymphocyte antigen 86 |
| chr7_+_50304693 | 4.41 |
ENST00000331340.8
ENST00000413698.5 ENST00000612658.4 ENST00000359197.9 ENST00000349824.8 ENST00000343574.9 ENST00000357364.8 ENST00000440768.6 ENST00000346667.8 ENST00000615491.4 |
IKZF1
|
IKAROS family zinc finger 1 |
| chr1_+_160739265 | 4.40 |
ENST00000368042.7
|
SLAMF7
|
SLAM family member 7 |
| chr2_-_89117844 | 4.35 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr21_+_29300770 | 4.35 |
ENST00000447177.5
|
BACH1
|
BTB domain and CNC homolog 1 |
| chr1_-_157700738 | 4.34 |
ENST00000368186.9
ENST00000496769.1 ENST00000368184.8 |
FCRL3
|
Fc receptor like 3 |
| chr2_+_90114838 | 4.33 |
ENST00000417279.3
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 |
| chr1_+_198639162 | 4.31 |
ENST00000418674.1
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr11_-_59212869 | 4.28 |
ENST00000361050.4
|
MPEG1
|
macrophage expressed 1 |
| chr14_-_106185387 | 4.27 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18 |
| chr1_+_160739239 | 3.97 |
ENST00000368043.8
|
SLAMF7
|
SLAM family member 7 |
| chr16_+_85902689 | 3.93 |
ENST00000563180.1
ENST00000564617.5 ENST00000564803.5 |
IRF8
|
interferon regulatory factor 8 |
| chr14_+_22007503 | 3.91 |
ENST00000390447.3
|
TRAV19
|
T cell receptor alpha variable 19 |
| chr14_-_106511856 | 3.90 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr14_-_106062670 | 3.82 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr7_-_36724457 | 3.77 |
ENST00000617537.5
ENST00000435386.1 |
AOAH
|
acyloxyacyl hydrolase |
| chr3_+_111542134 | 3.75 |
ENST00000438817.6
|
CD96
|
CD96 molecule |
| chr12_-_10130143 | 3.74 |
ENST00000298523.9
ENST00000396484.6 ENST00000310002.4 ENST00000304084.13 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr14_+_21712313 | 3.73 |
ENST00000390424.2
|
TRAV2
|
T cell receptor alpha variable 2 |
| chr14_+_22112280 | 3.68 |
ENST00000390454.2
|
TRAV25
|
T cell receptor alpha variable 25 |
| chr2_+_230225718 | 3.67 |
ENST00000420434.7
ENST00000392045.8 ENST00000417495.7 ENST00000343805.10 |
SP140
|
SP140 nuclear body protein |
| chr14_-_95714114 | 3.66 |
ENST00000402399.6
ENST00000555202.1 |
TCL1A
|
TCL1 family AKT coactivator A |
| chr1_-_206921867 | 3.65 |
ENST00000628511.2
ENST00000367091.8 |
FCMR
|
Fc fragment of IgM receptor |
| chr12_+_69348372 | 3.62 |
ENST00000261267.7
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chr1_+_116754422 | 3.59 |
ENST00000369478.4
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr7_+_139829153 | 3.58 |
ENST00000652056.1
|
TBXAS1
|
thromboxane A synthase 1 |
| chr22_+_22811737 | 3.56 |
ENST00000390315.3
|
IGLV3-10
|
immunoglobulin lambda variable 3-10 |
| chr14_-_106088573 | 3.54 |
ENST00000632099.1
|
IGHV3-64D
|
immunoglobulin heavy variable 3-64D |
| chr14_+_21749163 | 3.54 |
ENST00000390427.3
|
TRAV5
|
T cell receptor alpha variable 5 |
| chr14_-_106422175 | 3.51 |
ENST00000390619.2
|
IGHV4-39
|
immunoglobulin heavy variable 4-39 |
| chr14_-_106360320 | 3.48 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
| chrX_+_136648138 | 3.47 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand |
| chr11_+_65879791 | 3.46 |
ENST00000528419.6
ENST00000307886.8 ENST00000526034.2 ENST00000679584.1 ENST00000680443.1 ENST00000680670.1 |
CTSW
|
cathepsin W |
| chr22_+_22906342 | 3.46 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr11_-_118225086 | 3.45 |
ENST00000640745.1
|
JAML
|
junction adhesion molecule like |
| chr6_-_25042003 | 3.39 |
ENST00000510784.8
|
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr14_-_106771020 | 3.38 |
ENST00000617374.2
|
IGHV2-70
|
immunoglobulin heavy variable 2-70 |
| chr2_+_90172802 | 3.38 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr3_+_111542178 | 3.36 |
ENST00000283285.10
ENST00000352690.9 |
CD96
|
CD96 molecule |
| chr14_-_106811131 | 3.36 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr14_-_106154113 | 3.34 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr12_-_10130241 | 3.33 |
ENST00000353231.9
ENST00000525605.1 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr4_-_48080172 | 3.33 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr19_+_54593619 | 3.32 |
ENST00000251372.8
ENST00000453777.1 |
LILRA1
|
leukocyte immunoglobulin like receptor A1 |
| chr19_-_54281145 | 3.32 |
ENST00000434421.5
ENST00000391749.4 |
LILRB2
|
leukocyte immunoglobulin like receptor B2 |
| chr22_+_44172932 | 3.25 |
ENST00000422871.5
|
PARVG
|
parvin gamma |
| chrX_+_124346525 | 3.24 |
ENST00000360027.4
|
SH2D1A
|
SH2 domain containing 1A |
| chr7_+_142529268 | 3.23 |
ENST00000612787.1
|
TRBV7-9
|
T cell receptor beta variable 7-9 |
| chr14_-_95714146 | 3.22 |
ENST00000554012.5
|
TCL1A
|
TCL1 family AKT coactivator A |
| chr14_+_21924033 | 3.17 |
ENST00000390440.2
|
TRAV14DV4
|
T cell receptor alpha variable 14/delta variable 4 |
| chr12_+_10307950 | 3.17 |
ENST00000543420.5
ENST00000543777.5 |
KLRD1
|
killer cell lectin like receptor D1 |
| chr14_-_106737547 | 3.15 |
ENST00000632209.1
|
IGHV1-69-2
|
immunoglobulin heavy variable 1-69-2 |
| chr19_-_53824288 | 3.15 |
ENST00000324134.11
ENST00000391773.6 ENST00000391775.7 ENST00000345770.9 ENST00000391772.1 |
NLRP12
|
NLR family pyrin domain containing 12 |
| chrX_+_136648214 | 3.13 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr4_-_70666492 | 3.10 |
ENST00000254801.9
ENST00000391614.7 |
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr5_+_157180816 | 3.08 |
ENST00000422843.8
|
ITK
|
IL2 inducible T cell kinase |
| chr4_-_38856807 | 3.05 |
ENST00000506146.5
ENST00000436693.6 ENST00000508254.5 ENST00000514655.1 |
TLR1
TLR6
|
toll like receptor 1 toll like receptor 6 |
| chr7_+_139829242 | 3.04 |
ENST00000455353.6
ENST00000458722.6 ENST00000448866.7 ENST00000411653.6 |
TBXAS1
|
thromboxane A synthase 1 |
| chr20_+_59019397 | 3.03 |
ENST00000217133.2
|
TUBB1
|
tubulin beta 1 class VI |
| chr1_+_160739286 | 2.98 |
ENST00000359331.8
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr4_+_101813810 | 2.97 |
ENST00000444316.2
|
BANK1
|
B cell scaffold protein with ankyrin repeats 1 |
| chr7_+_142720652 | 2.96 |
ENST00000390400.2
|
TRBV28
|
T cell receptor beta variable 28 |
| chr15_-_79971164 | 2.95 |
ENST00000335661.6
ENST00000267953.4 ENST00000677151.1 |
BCL2A1
|
BCL2 related protein A1 |
| chr7_+_37683847 | 2.95 |
ENST00000447769.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr4_-_70666884 | 2.91 |
ENST00000510614.5
|
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr12_+_10307818 | 2.88 |
ENST00000350274.9
ENST00000336164.9 |
KLRD1
|
killer cell lectin like receptor D1 |
| chr13_-_99258366 | 2.87 |
ENST00000397470.5
ENST00000397473.7 |
GPR18
|
G protein-coupled receptor 18 |
| chr11_-_118212885 | 2.87 |
ENST00000524477.5
|
JAML
|
junction adhesion molecule like |
| chr14_-_106803221 | 2.86 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr14_-_95714088 | 2.84 |
ENST00000556450.5
|
TCL1A
|
TCL1 family AKT coactivator A |
| chr7_+_142740206 | 2.83 |
ENST00000422143.2
|
TRBV29-1
|
T cell receptor beta variable 29-1 |
| chr1_+_207496229 | 2.82 |
ENST00000367051.6
ENST00000367053.6 ENST00000367052.6 |
CR1
|
complement C3b/C4b receptor 1 (Knops blood group) |
| chr2_+_89959979 | 2.82 |
ENST00000453166.2
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr21_+_38256698 | 2.81 |
ENST00000613499.4
ENST00000612702.4 ENST00000398925.5 ENST00000398928.5 ENST00000328656.8 ENST00000443341.5 |
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr1_-_206921987 | 2.81 |
ENST00000530505.1
ENST00000442471.4 |
FCMR
|
Fc fragment of IgM receptor |
| chr14_-_106658251 | 2.76 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64 |
| chr7_-_36724380 | 2.73 |
ENST00000617267.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr4_-_70666961 | 2.71 |
ENST00000510437.5
|
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr1_-_157820113 | 2.68 |
ENST00000368176.8
|
FCRL1
|
Fc receptor like 1 |
| chr1_-_162412117 | 2.67 |
ENST00000367929.3
|
SH2D1B
|
SH2 domain containing 1B |
| chr7_-_38300288 | 2.65 |
ENST00000390341.2
|
TRGV10
|
T cell receptor gamma variable 10 (non-functional) |
| chr2_+_90159840 | 2.64 |
ENST00000377032.5
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr14_-_24609660 | 2.64 |
ENST00000557220.6
ENST00000216338.9 ENST00000382548.4 |
GZMH
|
granzyme H |
| chrX_+_129779930 | 2.61 |
ENST00000356892.4
|
SASH3
|
SAM and SH3 domain containing 3 |
| chr22_+_38952718 | 2.61 |
ENST00000402255.5
|
APOBEC3A
|
apolipoprotein B mRNA editing enzyme catalytic subunit 3A |
| chr12_+_31962466 | 2.60 |
ENST00000381054.3
|
RESF1
|
retroelement silencing factor 1 |
| chr2_-_89222461 | 2.59 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28 |
| chr14_-_106579223 | 2.58 |
ENST00000390626.2
|
IGHV5-51
|
immunoglobulin heavy variable 5-51 |
| chr1_-_150765735 | 2.55 |
ENST00000679898.1
ENST00000448301.7 ENST00000680664.1 ENST00000679512.1 ENST00000368985.8 ENST00000679582.1 |
CTSS
|
cathepsin S |
| chr12_-_10454485 | 2.54 |
ENST00000408006.7
ENST00000544822.2 ENST00000536188.5 |
KLRC1
|
killer cell lectin like receptor C1 |
| chr3_+_122325237 | 2.51 |
ENST00000264474.4
ENST00000479204.1 |
CSTA
|
cystatin A |
| chr14_-_106130061 | 2.50 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr21_+_42403874 | 2.49 |
ENST00000319294.11
ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr18_+_24113341 | 2.47 |
ENST00000540918.2
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr5_+_55024250 | 2.46 |
ENST00000231009.3
|
GZMK
|
granzyme K |
| chr22_-_17199609 | 2.45 |
ENST00000330232.8
|
ADA2
|
adenosine deaminase 2 |
| chr14_+_20891385 | 2.45 |
ENST00000304639.4
|
RNASE3
|
ribonuclease A family member 3 |
| chr2_-_89085787 | 2.44 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr2_+_230225756 | 2.43 |
ENST00000373645.3
|
SP140
|
SP140 nuclear body protein |
| chr1_+_158845798 | 2.42 |
ENST00000438394.1
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr14_+_22040576 | 2.40 |
ENST00000390448.3
|
TRAV20
|
T cell receptor alpha variable 20 |
| chr15_+_58138368 | 2.40 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chr21_+_38256984 | 2.40 |
ENST00000398938.7
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr14_-_106627685 | 2.36 |
ENST00000390629.3
|
IGHV4-59
|
immunoglobulin heavy variable 4-59 |
| chr12_-_10130082 | 2.36 |
ENST00000533022.5
|
CLEC7A
|
C-type lectin domain containing 7A |
| chr1_+_207496147 | 2.34 |
ENST00000400960.7
ENST00000367049.9 |
CR1
|
complement C3b/C4b receptor 1 (Knops blood group) |
| chr3_+_155083523 | 2.33 |
ENST00000680057.1
|
MME
|
membrane metalloendopeptidase |
| chr13_-_46182136 | 2.33 |
ENST00000323076.7
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr1_+_26529745 | 2.33 |
ENST00000374168.7
ENST00000374166.8 |
RPS6KA1
|
ribosomal protein S6 kinase A1 |
| chr4_+_73853290 | 2.31 |
ENST00000226524.4
|
PF4V1
|
platelet factor 4 variant 1 |
| chr12_+_47216531 | 2.31 |
ENST00000548348.1
ENST00000549500.1 |
PCED1B
|
PC-esterase domain containing 1B |
| chr2_+_89936859 | 2.29 |
ENST00000474213.1
|
IGKV2D-30
|
immunoglobulin kappa variable 2D-30 |
| chr14_+_21846534 | 2.29 |
ENST00000390434.3
|
TRAV8-2
|
T cell receptor alpha variable 8-2 |
| chr20_-_36646146 | 2.27 |
ENST00000262866.9
|
SLA2
|
Src like adaptor 2 |
| chr21_+_29300111 | 2.25 |
ENST00000451655.5
|
BACH1
|
BTB domain and CNC homolog 1 |
| chr7_+_142492121 | 2.24 |
ENST00000390374.3
|
TRBV7-6
|
T cell receptor beta variable 7-6 |
| chr19_-_51750798 | 2.23 |
ENST00000600815.1
|
FPR1
|
formyl peptide receptor 1 |
| chr17_-_78128630 | 2.23 |
ENST00000306591.11
|
TMC6
|
transmembrane channel like 6 |
| chr2_+_90021567 | 2.23 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr1_+_203765168 | 2.23 |
ENST00000367217.5
ENST00000442561.7 |
LAX1
|
lymphocyte transmembrane adaptor 1 |
| chr14_+_22070548 | 2.22 |
ENST00000390450.3
|
TRAV22
|
T cell receptor alpha variable 22 |
| chr4_+_40192949 | 2.22 |
ENST00000507851.5
ENST00000615577.4 ENST00000613272.4 |
RHOH
|
ras homolog family member H |
| chr5_+_35856883 | 2.19 |
ENST00000506850.5
ENST00000303115.8 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr14_+_22163226 | 2.18 |
ENST00000390458.3
|
TRAV29DV5
|
T cell receptor alpha variable 29/delta variable 5 |
| chr22_+_36913620 | 2.18 |
ENST00000403662.8
ENST00000262825.9 |
CSF2RB
|
colony stimulating factor 2 receptor subunit beta |
| chr3_+_155083889 | 2.16 |
ENST00000680282.1
|
MME
|
membrane metalloendopeptidase |
| chr14_+_21965451 | 2.16 |
ENST00000390442.3
|
TRAV12-3
|
T cell receptor alpha variable 12-3 |
| chr14_+_21997531 | 2.15 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr22_+_35383106 | 2.14 |
ENST00000678411.1
|
HMOX1
|
heme oxygenase 1 |
| chr5_+_76609091 | 2.13 |
ENST00000514001.5
ENST00000396234.7 ENST00000509074.5 ENST00000502745.5 |
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr7_+_74777269 | 2.13 |
ENST00000442021.6
ENST00000433458.5 |
NCF1
|
neutrophil cytosolic factor 1 |
| chrY_+_2841594 | 2.13 |
ENST00000250784.13
|
RPS4Y1
|
ribosomal protein S4 Y-linked 1 |
| chr2_-_89268506 | 2.13 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chrX_+_124346571 | 2.12 |
ENST00000477673.2
|
SH2D1A
|
SH2 domain containing 1A |
| chr7_-_36724543 | 2.11 |
ENST00000612871.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr1_-_113871665 | 2.10 |
ENST00000528414.5
ENST00000460620.5 ENST00000359785.10 ENST00000420377.6 ENST00000525799.1 ENST00000538253.5 |
PTPN22
|
protein tyrosine phosphatase non-receptor type 22 |
| chr15_-_55270383 | 2.10 |
ENST00000396307.6
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr7_-_38330935 | 2.08 |
ENST00000390343.2
|
TRGV8
|
T cell receptor gamma variable 8 |
| chr3_-_3110347 | 2.08 |
ENST00000383846.5
ENST00000427088.1 ENST00000446632.7 ENST00000418488.6 ENST00000438560.5 |
IL5RA
|
interleukin 5 receptor subunit alpha |
| chr11_+_4449295 | 2.06 |
ENST00000325719.4
|
OR52K2
|
olfactory receptor family 52 subfamily K member 2 |
| chr12_+_75481204 | 2.05 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr1_+_207496268 | 2.05 |
ENST00000529814.1
|
CR1
|
complement C3b/C4b receptor 1 (Knops blood group) |
| chr2_+_90004792 | 2.04 |
ENST00000462693.1
|
IGKV2D-24
|
immunoglobulin kappa variable 2D-24 (non-functional) |
| chr6_-_127918604 | 2.04 |
ENST00000537166.5
|
THEMIS
|
thymocyte selection associated |
| chr12_+_40310431 | 2.03 |
ENST00000681696.1
|
LRRK2
|
leucine rich repeat kinase 2 |
| chr2_-_88947820 | 2.02 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr17_-_78128731 | 2.02 |
ENST00000592063.5
ENST00000590602.6 ENST00000589271.5 |
TMC6
|
transmembrane channel like 6 |
| chr14_-_105626066 | 2.01 |
ENST00000641978.1
ENST00000390543.3 |
IGHG4
|
immunoglobulin heavy constant gamma 4 (G4m marker) |
| chr1_-_160579439 | 2.01 |
ENST00000368054.8
ENST00000368048.7 ENST00000311224.8 ENST00000368051.3 ENST00000534968.5 |
CD84
|
CD84 molecule |
| chr1_-_159076742 | 1.99 |
ENST00000368130.9
|
AIM2
|
absent in melanoma 2 |
| chr14_+_22226711 | 1.98 |
ENST00000390463.3
|
TRAV36DV7
|
T cell receptor alpha variable 36/delta variable 7 |
| chr19_+_54874275 | 1.98 |
ENST00000469767.5
ENST00000391725.7 ENST00000345937.8 ENST00000353758.8 ENST00000359272.8 ENST00000391723.7 ENST00000391724.3 |
FCAR
|
Fc fragment of IgA receptor |
| chr1_-_150765785 | 1.97 |
ENST00000680311.1
ENST00000681728.1 ENST00000680288.1 |
CTSS
|
cathepsin S |
| chr1_-_89126066 | 1.95 |
ENST00000370466.4
|
GBP2
|
guanylate binding protein 2 |
| chr19_+_35902486 | 1.95 |
ENST00000246551.9
ENST00000437550.2 |
HCST
|
hematopoietic cell signal transducer |
| chr6_+_33075952 | 1.94 |
ENST00000418931.7
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr10_-_72088972 | 1.94 |
ENST00000317376.8
ENST00000412663.5 |
SPOCK2
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 2 |
| chr15_-_55249029 | 1.91 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr15_-_55270874 | 1.91 |
ENST00000567380.5
ENST00000565972.5 ENST00000569493.5 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr14_-_106791226 | 1.91 |
ENST00000433072.2
|
IGHV3-72
|
immunoglobulin heavy variable 3-72 |
| chr1_+_40396766 | 1.89 |
ENST00000539317.2
|
SMAP2
|
small ArfGAP2 |
| chr12_-_10409757 | 1.89 |
ENST00000309384.2
|
KLRC4
|
killer cell lectin like receptor C4 |
| chr1_-_157820060 | 1.87 |
ENST00000491942.1
ENST00000358292.7 |
FCRL1
|
Fc receptor like 1 |
| chr9_-_120914549 | 1.87 |
ENST00000546084.5
|
TRAF1
|
TNF receptor associated factor 1 |
| chr18_+_63897152 | 1.86 |
ENST00000397996.6
ENST00000418725.1 |
SERPINB10
|
serpin family B member 10 |
| chr12_-_54981838 | 1.85 |
ENST00000316577.12
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr12_-_110445540 | 1.85 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex subunit 3 |
| chr19_-_23687163 | 1.84 |
ENST00000601010.5
ENST00000601935.5 ENST00000600313.5 ENST00000596211.5 ENST00000359788.9 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr6_-_111759910 | 1.83 |
ENST00000517419.5
|
FYN
|
FYN proto-oncogene, Src family tyrosine kinase |
| chr16_+_50696999 | 1.82 |
ENST00000300589.6
|
NOD2
|
nucleotide binding oligomerization domain containing 2 |
| chr6_-_127918586 | 1.81 |
ENST00000626040.2
|
THEMIS
|
thymocyte selection associated |
| chr14_-_106277039 | 1.79 |
ENST00000390610.2
|
IGHV1-24
|
immunoglobulin heavy variable 1-24 |
| chr2_-_230225628 | 1.77 |
ENST00000540870.5
|
SP110
|
SP110 nuclear body protein |
| chrX_+_41689006 | 1.77 |
ENST00000378138.5
ENST00000620846.1 ENST00000649219.1 |
GPR34
|
G protein-coupled receptor 34 |
| chr14_+_21825453 | 1.77 |
ENST00000390432.2
|
TRAV10
|
T cell receptor alpha variable 10 |
| chrX_+_12975083 | 1.76 |
ENST00000451311.7
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4 X-linked |
| chr14_+_22105305 | 1.76 |
ENST00000390453.1
|
TRAV24
|
T cell receptor alpha variable 24 |
| chr15_+_74788542 | 1.74 |
ENST00000567571.5
|
CSK
|
C-terminal Src kinase |
| chr6_+_167122742 | 1.74 |
ENST00000341935.9
ENST00000349984.6 |
CCR6
|
C-C motif chemokine receptor 6 |
| chr16_+_21612637 | 1.74 |
ENST00000568826.1
|
METTL9
|
methyltransferase like 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFK
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 28.7 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 2.4 | 7.2 | GO:0030451 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 2.4 | 7.1 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 1.6 | 4.8 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 1.5 | 4.5 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 1.2 | 3.5 | GO:0034130 | toll-like receptor 1 signaling pathway(GO:0034130) |
| 1.2 | 4.6 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 1.1 | 6.6 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 1.0 | 5.1 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 1.0 | 8.1 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.9 | 12.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.9 | 6.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.9 | 5.2 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.8 | 6.3 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.8 | 8.7 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.8 | 3.9 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.7 | 123.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.7 | 2.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.7 | 2.0 | GO:1903123 | regulation of thioredoxin peroxidase activity(GO:1903123) negative regulation of thioredoxin peroxidase activity(GO:1903124) negative regulation of thioredoxin peroxidase activity by peptidyl-threonine phosphorylation(GO:1903125) Wnt signalosome assembly(GO:1904887) negative regulation of peroxidase activity(GO:2000469) |
| 0.6 | 0.6 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.6 | 3.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.6 | 3.4 | GO:2001107 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.5 | 3.3 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.5 | 8.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.5 | 10.1 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.5 | 1.6 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.5 | 1.6 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.5 | 4.1 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.5 | 1.5 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.5 | 2.4 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.5 | 2.4 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.5 | 14.3 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.5 | 4.5 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.4 | 3.6 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.4 | 2.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.4 | 2.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.4 | 5.5 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.4 | 3.0 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.4 | 3.4 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.4 | 14.4 | GO:0002717 | positive regulation of natural killer cell mediated immunity(GO:0002717) |
| 0.4 | 3.3 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.4 | 2.9 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.4 | 0.8 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.4 | 9.6 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.4 | 1.2 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.4 | 1.2 | GO:0002372 | myeloid dendritic cell cytokine production(GO:0002372) |
| 0.4 | 1.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.4 | 3.0 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.4 | 1.1 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.4 | 0.7 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.4 | 2.5 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.3 | 1.4 | GO:0071725 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.3 | 4.1 | GO:0038124 | toll-like receptor TLR6:TLR2 signaling pathway(GO:0038124) response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.3 | 2.0 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.3 | 3.0 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.3 | 2.6 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.3 | 1.6 | GO:0052148 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.3 | 1.2 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.3 | 2.2 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.3 | 0.6 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.3 | 2.8 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.3 | 0.9 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.3 | 6.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 1.8 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.3 | 2.0 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.3 | 1.4 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.3 | 16.4 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.3 | 2.3 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.3 | 5.5 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.3 | 2.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.3 | 0.8 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.2 | 1.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 1.5 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.2 | 2.1 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.2 | 2.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.2 | 2.3 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.2 | 0.9 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.2 | 2.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 3.3 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.2 | 3.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.2 | 0.7 | GO:1990579 | protein trans-autophosphorylation(GO:0036290) peptidyl-serine trans-autophosphorylation(GO:1990579) |
| 0.2 | 0.6 | GO:1901804 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.2 | 3.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 | 1.1 | GO:1904764 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.2 | 0.6 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.2 | 0.8 | GO:0034148 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.2 | 0.6 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 0.6 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.2 | 0.8 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.2 | 2.4 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 3.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 0.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.2 | 0.6 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.2 | 1.5 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.2 | 35.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.2 | 1.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 0.5 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.2 | 1.0 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.2 | 1.4 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 | 3.7 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.2 | 1.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 2.0 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.2 | 0.6 | GO:0060661 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.2 | 0.6 | GO:0033320 | UDP-D-xylose metabolic process(GO:0033319) UDP-D-xylose biosynthetic process(GO:0033320) |
| 0.1 | 0.9 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 | 0.7 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 | 0.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 1.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 1.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 12.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.4 | GO:0099403 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.1 | 1.9 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 1.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 3.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.7 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 1.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.9 | GO:0014900 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.1 | 0.5 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 1.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.6 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 | 0.5 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.1 | 1.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 1.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 5.6 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 1.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.2 | GO:0038156 | interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.1 | 0.8 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.1 | 2.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.6 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.8 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.1 | 0.2 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.1 | 2.3 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.8 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.4 | GO:2000699 | regulation of cell proliferation involved in mesonephros development(GO:2000606) negative regulation of cell proliferation involved in mesonephros development(GO:2000607) fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000699) glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000701) regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000702) negative regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000703) regulation of glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000733) negative regulation of glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000734) |
| 0.1 | 1.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.6 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.3 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.1 | 1.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 3.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.5 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.1 | 1.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 2.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.7 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 6.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 1.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 1.1 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.5 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.4 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 0.1 | GO:1901536 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 2.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.9 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.1 | 1.2 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.1 | 0.3 | GO:0002604 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.1 | 0.7 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.4 | GO:1904468 | negative regulation of tumor necrosis factor secretion(GO:1904468) |
| 0.1 | 0.3 | GO:0014810 | positive regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014810) |
| 0.1 | 2.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.9 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.3 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.3 | GO:1901874 | negative regulation of post-translational protein modification(GO:1901874) |
| 0.1 | 2.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 0.5 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.8 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 1.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.8 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.2 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.1 | 0.6 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.1 | 1.0 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.1 | 0.6 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 0.7 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.8 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.1 | 0.5 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 8.0 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 0.2 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.1 | 1.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 1.1 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.1 | 0.5 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.1 | 1.0 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.1 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.1 | 0.8 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.4 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.6 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.5 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.1 | 0.5 | GO:0072011 | glomerular endothelium development(GO:0072011) |
| 0.1 | 1.7 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 0.7 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 2.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.9 | GO:0014870 | response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.4 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.4 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 5.6 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.1 | 1.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.2 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.1 | 1.7 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 0.3 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.6 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.6 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.1 | 1.5 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 1.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.4 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 1.0 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.1 | 0.5 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 0.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.9 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.2 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.1 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.2 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 0.8 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.5 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 8.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.5 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.9 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.9 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 0.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 2.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.3 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.5 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.9 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 1.0 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 1.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.4 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.3 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) |
| 0.0 | 0.4 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.5 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.6 | GO:2000347 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) positive regulation of hepatocyte proliferation(GO:2000347) |
| 0.0 | 3.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.4 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 1.3 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.4 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.2 | GO:2000330 | positive regulation of activation of Janus kinase activity(GO:0010536) interleukin-23-mediated signaling pathway(GO:0038155) positive regulation of memory T cell differentiation(GO:0043382) regulation of NK T cell activation(GO:0051133) positive regulation of NK T cell activation(GO:0051135) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.6 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.0 | 2.9 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.6 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.4 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.9 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.2 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.4 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.7 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 1.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 1.1 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 3.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.3 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 1.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 8.6 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.2 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 1.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.2 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 1.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.4 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 1.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.3 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.4 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 4.3 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 0.1 | GO:1900920 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.0 | 0.9 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.2 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.4 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.6 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.4 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.3 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.5 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 1.6 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.0 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 1.0 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.2 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.3 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 1.8 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.4 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.9 | GO:0006625 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.2 | GO:0072093 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.7 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 1.0 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) |
| 0.0 | 0.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.9 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.0 | 3.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.8 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.1 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.0 | 0.2 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.6 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) |
| 0.0 | 0.7 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 2.5 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.7 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.3 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.9 | GO:0050715 | positive regulation of cytokine secretion(GO:0050715) |
| 0.0 | 0.4 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 3.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 1.1 | GO:0007077 | mitotic nuclear envelope disassembly(GO:0007077) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 1.1 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.9 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.3 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.4 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.1 | GO:0046070 | dGTP metabolic process(GO:0046070) |
| 0.0 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.5 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.2 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.3 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.0 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.7 | GO:0071757 | hexameric IgM immunoglobulin complex(GO:0071757) |
| 0.9 | 3.5 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.8 | 43.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.7 | 2.2 | GO:0030526 | granulocyte macrophage colony-stimulating factor receptor complex(GO:0030526) |
| 0.7 | 2.0 | GO:0032473 | cytoplasmic side of mitochondrial outer membrane(GO:0032473) caveola neck(GO:0099400) |
| 0.6 | 2.4 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.5 | 3.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.4 | 1.3 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.4 | 3.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 1.2 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.4 | 1.1 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.3 | 2.0 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.3 | 6.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.3 | 6.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 1.2 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.3 | 3.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.3 | 2.8 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.3 | 3.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.3 | 2.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 0.5 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.2 | 0.7 | GO:0090651 | apical cytoplasm(GO:0090651) |
| 0.2 | 0.9 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.2 | 0.6 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.2 | 1.0 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.2 | 26.1 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.2 | 39.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 1.2 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.2 | 4.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.2 | 2.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 25.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.2 | 3.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 1.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 12.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 8.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 57.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.5 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 2.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.5 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 6.6 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 2.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.7 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 0.5 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.1 | 3.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.8 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.6 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 8.6 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 0.3 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 1.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 0.5 | GO:0001652 | dense fibrillar component(GO:0001651) granular component(GO:0001652) |
| 0.1 | 1.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 1.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 2.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 1.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 6.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.5 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 0.4 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.5 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 4.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.9 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 2.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.5 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 5.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 3.6 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.6 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 1.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.4 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 1.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 1.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.7 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.2 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 1.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.5 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 2.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 3.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 4.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.1 | GO:0030892 | mitotic cohesin complex(GO:0030892) |
| 0.0 | 0.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 2.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.8 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 2.4 | 7.2 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 2.2 | 6.6 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 2.2 | 6.6 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 1.8 | 14.2 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 1.7 | 11.8 | GO:0019862 | IgA binding(GO:0019862) |
| 1.3 | 8.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 1.2 | 4.6 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 1.0 | 5.1 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.9 | 3.5 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.9 | 4.3 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.8 | 2.5 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.7 | 2.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.7 | 5.6 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.7 | 2.0 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) peroxidase inhibitor activity(GO:0036479) |
| 0.6 | 43.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.6 | 24.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.6 | 3.5 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.5 | 2.7 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.5 | 111.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.5 | 2.1 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.5 | 2.4 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.4 | 2.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.4 | 1.2 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.4 | 5.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.4 | 1.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.4 | 1.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.4 | 14.3 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.3 | 2.8 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.3 | 1.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.3 | 2.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.3 | 6.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.3 | 2.6 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.3 | 1.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 1.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 1.8 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 0.9 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 5.0 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 5.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.2 | 0.9 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.2 | 0.9 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.2 | 2.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 0.8 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 9.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 1.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 0.6 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.2 | 0.8 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.2 | 0.6 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.2 | 2.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.2 | 1.4 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.2 | 3.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 0.9 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 1.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.2 | 2.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 0.7 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.2 | 3.7 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 5.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 0.5 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.2 | 4.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 1.6 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 0.6 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.2 | 2.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 2.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 0.6 | GO:0048040 | UDP-glucuronate decarboxylase activity(GO:0048040) |
| 0.1 | 3.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.9 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 3.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.4 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.1 | 1.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.4 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.1 | 0.4 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 1.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 1.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.4 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 2.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.7 | GO:0017089 | glycolipid transporter activity(GO:0017089) ceramide binding(GO:0097001) |
| 0.1 | 0.6 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.4 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.1 | 0.6 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 1.0 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.4 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.1 | 0.4 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 0.8 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.2 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) |
| 0.1 | 0.3 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 2.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 2.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 1.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 9.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.6 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 2.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.1 | 3.1 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 1.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.8 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 2.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 2.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.8 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 3.0 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.5 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 0.6 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 1.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 3.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 2.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.2 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.1 | 2.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.2 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 1.0 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 1.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 1.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 4.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.2 | GO:0090422 | thiamine pyrophosphate transporter activity(GO:0090422) |
| 0.1 | 0.3 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 3.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.2 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 0.2 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.8 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.1 | 0.7 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.1 | 0.8 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 1.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.4 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.5 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.4 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 0.3 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 1.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.2 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 0.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.6 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 6.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 1.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.7 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.6 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.0 | 2.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.4 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 1.3 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 2.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.7 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.9 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 2.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 1.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.7 | GO:0031701 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 1.0 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0051990 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.0 | 0.1 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 0.7 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 1.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 2.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 1.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 7.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 8.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.5 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 2.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 3.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.0 | 1.0 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.0 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 2.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0016416 | O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 1.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.0 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 1.0 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.1 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 1.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 2.1 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 31.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.2 | 1.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 6.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 5.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 10.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 5.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 4.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 5.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 6.3 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 5.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 10.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 2.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 5.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 2.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 5.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.9 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 4.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 4.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 1.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 3.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 2.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 3.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.7 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 5.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.9 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.4 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 33.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.4 | 4.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.4 | 43.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 1.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 3.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 5.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 6.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 5.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 2.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 9.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 3.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 2.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 4.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 7.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 5.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 6.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 12.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 3.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 2.9 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 3.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.9 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 0.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 6.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 3.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.5 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 6.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.9 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.1 | 1.5 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.1 | 1.8 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |
| 0.1 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 0.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 3.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.5 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.8 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 2.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 2.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 3.2 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.2 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 2.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 2.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 2.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.8 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 5.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.5 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |