Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for MAF_NRL
Z-value: 1.66
Transcription factors associated with MAF_NRL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
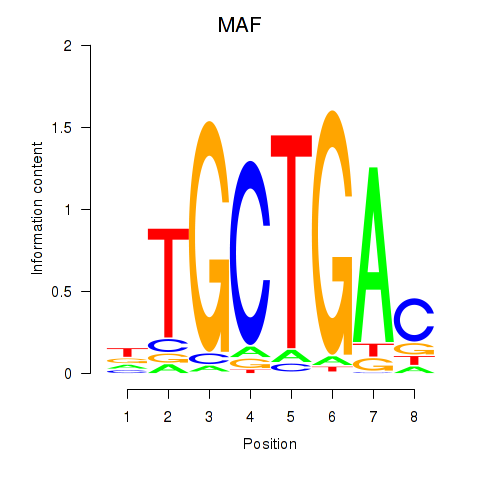
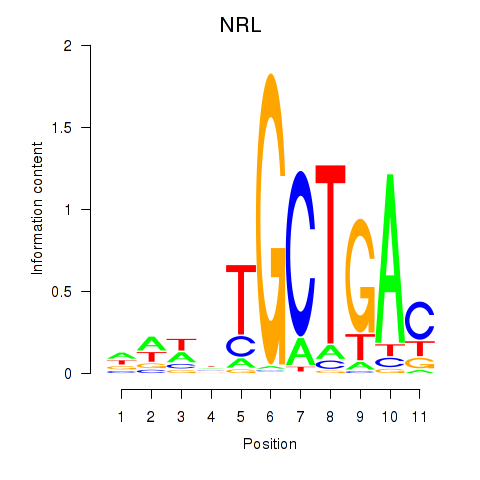
MAF
|
ENSG00000178573.7 | MAF bZIP transcription factor |
|
NRL
|
ENSG00000129535.13 | neural retina leucine zipper |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NRL | hg38_v1_chr14_-_24084625_24084641 | -0.26 | 1.6e-01 | Click! |
| MAF | hg38_v1_chr16_-_79600698_79600723 | 0.09 | 6.3e-01 | Click! |
Activity profile of MAF_NRL motif
Sorted Z-values of MAF_NRL motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_116829898 | 11.38 |
ENST00000227667.8
ENST00000375345.3 |
APOC3
|
apolipoprotein C3 |
| chr4_+_77605807 | 10.63 |
ENST00000682537.1
|
CXCL13
|
C-X-C motif chemokine ligand 13 |
| chr1_-_56966133 | 10.61 |
ENST00000535057.5
ENST00000543257.5 |
C8B
|
complement C8 beta chain |
| chr1_-_56966006 | 9.97 |
ENST00000371237.9
|
C8B
|
complement C8 beta chain |
| chr11_+_116830529 | 8.45 |
ENST00000630701.1
|
APOC3
|
apolipoprotein C3 |
| chr22_+_20774092 | 7.50 |
ENST00000215727.10
|
SERPIND1
|
serpin family D member 1 |
| chr1_+_207104226 | 6.07 |
ENST00000367070.8
|
C4BPA
|
complement component 4 binding protein alpha |
| chr1_+_207104287 | 5.99 |
ENST00000421786.5
|
C4BPA
|
complement component 4 binding protein alpha |
| chr5_-_156963222 | 5.80 |
ENST00000407087.4
ENST00000274532.7 |
TIMD4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr19_+_44914247 | 5.51 |
ENST00000588750.5
ENST00000588802.5 |
APOC1
|
apolipoprotein C1 |
| chr9_-_114078293 | 5.45 |
ENST00000265132.8
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr14_-_105771405 | 5.02 |
ENST00000641136.1
ENST00000390551.6 |
IGHG3
|
immunoglobulin heavy constant gamma 3 (G3m marker) |
| chr19_+_44914833 | 4.82 |
ENST00000589078.1
ENST00000586638.5 |
APOC1
|
apolipoprotein C1 |
| chr9_+_136945234 | 4.53 |
ENST00000371634.7
|
C8G
|
complement C8 gamma chain |
| chr5_+_132873660 | 4.35 |
ENST00000296877.3
|
LEAP2
|
liver enriched antimicrobial peptide 2 |
| chr19_+_44914588 | 4.20 |
ENST00000592535.6
|
APOC1
|
apolipoprotein C1 |
| chr16_+_176659 | 4.16 |
ENST00000320868.9
ENST00000397797.1 |
HBA1
|
hemoglobin subunit alpha 1 |
| chr10_+_94683722 | 4.14 |
ENST00000285979.11
|
CYP2C18
|
cytochrome P450 family 2 subfamily C member 18 |
| chr22_+_22811737 | 4.04 |
ENST00000390315.3
|
IGLV3-10
|
immunoglobulin lambda variable 3-10 |
| chr2_-_187554351 | 3.70 |
ENST00000437725.5
ENST00000409676.5 ENST00000233156.9 ENST00000339091.8 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor |
| chr6_-_7910776 | 3.66 |
ENST00000379757.9
|
TXNDC5
|
thioredoxin domain containing 5 |
| chr14_-_106538331 | 3.64 |
ENST00000390624.3
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr11_-_116837586 | 3.63 |
ENST00000375320.5
ENST00000359492.6 ENST00000375329.6 ENST00000375323.5 ENST00000236850.5 |
APOA1
|
apolipoprotein A1 |
| chr17_+_42900791 | 3.55 |
ENST00000592383.5
ENST00000253801.7 ENST00000585489.1 |
G6PC1
|
glucose-6-phosphatase catalytic subunit 1 |
| chr22_+_22880706 | 3.52 |
ENST00000390319.2
|
IGLV3-1
|
immunoglobulin lambda variable 3-1 |
| chr10_+_94683771 | 3.45 |
ENST00000339022.6
|
CYP2C18
|
cytochrome P450 family 2 subfamily C member 18 |
| chr14_-_106235582 | 3.26 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr20_+_46709623 | 3.22 |
ENST00000359271.4
|
SLC2A10
|
solute carrier family 2 member 10 |
| chrX_-_106038721 | 3.22 |
ENST00000372563.2
|
SERPINA7
|
serpin family A member 7 |
| chr3_+_52794768 | 3.21 |
ENST00000621946.4
ENST00000416872.6 ENST00000449956.2 |
ITIH3
|
inter-alpha-trypsin inhibitor heavy chain 3 |
| chr11_+_78063851 | 2.93 |
ENST00000281030.2
|
THRSP
|
thyroid hormone responsive |
| chr12_-_52903648 | 2.85 |
ENST00000546900.1
|
KRT8
|
keratin 8 |
| chr6_+_31927683 | 2.82 |
ENST00000456570.5
|
ENSG00000244255.5
|
novel complement component 2 (C2) and complement factor B (CFB) protein |
| chr1_-_153608136 | 2.76 |
ENST00000368703.6
|
S100A16
|
S100 calcium binding protein A16 |
| chr2_-_187554473 | 2.73 |
ENST00000453013.5
ENST00000417013.5 |
TFPI
|
tissue factor pathway inhibitor |
| chr3_-_50303565 | 2.72 |
ENST00000266031.8
ENST00000395143.6 ENST00000457214.6 ENST00000447605.2 ENST00000395144.7 ENST00000418723.1 |
HYAL1
|
hyaluronidase 1 |
| chr2_+_88885397 | 2.67 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr6_+_31927703 | 2.63 |
ENST00000418949.6
ENST00000299367.10 ENST00000383177.7 ENST00000477310.1 |
C2
ENSG00000244255.5
|
complement C2 novel complement component 2 (C2) and complement factor B (CFB) protein |
| chr6_+_31927486 | 2.62 |
ENST00000442278.6
|
C2
|
complement C2 |
| chr17_+_6641008 | 2.51 |
ENST00000570330.5
|
TXNDC17
|
thioredoxin domain containing 17 |
| chr9_-_33447553 | 2.49 |
ENST00000645858.1
ENST00000297991.6 |
AQP3
|
aquaporin 3 (Gill blood group) |
| chr20_-_49482645 | 2.49 |
ENST00000371741.6
|
KCNB1
|
potassium voltage-gated channel subfamily B member 1 |
| chr22_+_22380766 | 2.49 |
ENST00000390297.3
|
IGLV1-44
|
immunoglobulin lambda variable 1-44 |
| chr3_+_186613052 | 2.43 |
ENST00000411641.7
ENST00000273784.5 |
AHSG
|
alpha 2-HS glycoprotein |
| chr1_-_157841800 | 2.31 |
ENST00000368174.5
|
CD5L
|
CD5 molecule like |
| chr5_+_42756811 | 2.30 |
ENST00000388827.4
ENST00000361970.10 |
CCDC152
|
coiled-coil domain containing 152 |
| chr15_-_100341899 | 2.25 |
ENST00000568565.2
ENST00000268070.9 |
ADAMTS17
|
ADAM metallopeptidase with thrombospondin type 1 motif 17 |
| chr12_-_102480552 | 2.15 |
ENST00000337514.11
ENST00000307046.8 |
IGF1
|
insulin like growth factor 1 |
| chr2_+_233671879 | 2.14 |
ENST00000354728.5
|
UGT1A9
|
UDP glucuronosyltransferase family 1 member A9 |
| chr6_+_32038382 | 2.14 |
ENST00000478281.5
ENST00000471671.4 ENST00000435122.3 ENST00000644719.2 |
CYP21A2
|
cytochrome P450 family 21 subfamily A member 2 |
| chr3_+_52779916 | 2.11 |
ENST00000537050.5
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr14_-_105743032 | 2.09 |
ENST00000390548.6
ENST00000390549.6 ENST00000390542.6 |
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker) |
| chr6_+_31927510 | 2.08 |
ENST00000447952.6
|
C2
|
complement C2 |
| chr11_-_10568650 | 2.07 |
ENST00000256178.8
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr11_-_10568571 | 2.05 |
ENST00000529598.1
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr12_-_102917114 | 2.04 |
ENST00000550978.6
|
PAH
|
phenylalanine hydroxylase |
| chr2_+_127423265 | 2.03 |
ENST00000402125.2
|
PROC
|
protein C, inactivator of coagulation factors Va and VIIIa |
| chr7_-_100641507 | 2.01 |
ENST00000431692.5
ENST00000223051.8 |
TFR2
|
transferrin receptor 2 |
| chr2_+_134838610 | 2.00 |
ENST00000356140.10
ENST00000392928.5 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr22_+_22409755 | 1.99 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr1_-_201399525 | 1.99 |
ENST00000367313.4
|
LAD1
|
ladinin 1 |
| chr9_+_113594118 | 1.97 |
ENST00000620489.1
|
RGS3
|
regulator of G protein signaling 3 |
| chr3_-_52826834 | 1.94 |
ENST00000441637.2
|
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain 4 |
| chr2_-_87825952 | 1.94 |
ENST00000398146.4
|
RGPD2
|
RANBP2 like and GRIP domain containing 2 |
| chr11_+_75151095 | 1.94 |
ENST00000289575.10
ENST00000525650.5 ENST00000454962.6 |
SLCO2B1
|
solute carrier organic anion transporter family member 2B1 |
| chr2_+_233691607 | 1.92 |
ENST00000373424.5
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase family 1 member A6 |
| chr2_-_89085787 | 1.92 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr9_+_27109393 | 1.91 |
ENST00000406359.8
|
TEK
|
TEK receptor tyrosine kinase |
| chr14_+_69398683 | 1.86 |
ENST00000556605.5
ENST00000031146.8 ENST00000336643.10 |
SLC39A9
|
solute carrier family 39 member 9 |
| chr12_+_103587266 | 1.83 |
ENST00000388887.7
|
STAB2
|
stabilin 2 |
| chr14_-_105940235 | 1.80 |
ENST00000390593.2
|
IGHV6-1
|
immunoglobulin heavy variable 6-1 |
| chr1_-_206946448 | 1.78 |
ENST00000356495.5
|
PIGR
|
polymeric immunoglobulin receptor |
| chr4_+_165378998 | 1.76 |
ENST00000402744.9
|
CPE
|
carboxypeptidase E |
| chr12_-_102480638 | 1.75 |
ENST00000392904.5
|
IGF1
|
insulin like growth factor 1 |
| chr18_+_24113341 | 1.72 |
ENST00000540918.2
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr2_+_119429889 | 1.72 |
ENST00000409826.1
ENST00000417645.1 |
TMEM37
|
transmembrane protein 37 |
| chr14_-_106771020 | 1.66 |
ENST00000617374.2
|
IGHV2-70
|
immunoglobulin heavy variable 2-70 |
| chr9_+_27109441 | 1.65 |
ENST00000519080.1
|
TEK
|
TEK receptor tyrosine kinase |
| chr2_+_113127588 | 1.65 |
ENST00000409930.4
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr12_+_118136086 | 1.61 |
ENST00000261313.3
|
PEBP1
|
phosphatidylethanolamine binding protein 1 |
| chr22_+_22431949 | 1.61 |
ENST00000390301.3
|
IGLV1-36
|
immunoglobulin lambda variable 1-36 |
| chr22_+_22357739 | 1.61 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr12_-_95996302 | 1.61 |
ENST00000261208.8
ENST00000538703.5 ENST00000541929.5 |
HAL
|
histidine ammonia-lyase |
| chr11_-_2903490 | 1.60 |
ENST00000455942.3
ENST00000625099.4 |
SLC22A18AS
|
solute carrier family 22 member 18 antisense |
| chr4_+_110476133 | 1.57 |
ENST00000265162.10
|
ENPEP
|
glutamyl aminopeptidase |
| chr2_-_88979016 | 1.57 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr14_-_22589157 | 1.57 |
ENST00000538631.1
ENST00000543337.1 ENST00000250498.9 |
DAD1
|
defender against cell death 1 |
| chr9_-_125241327 | 1.55 |
ENST00000324460.7
ENST00000680272.1 ENST00000680032.1 |
HSPA5
|
heat shock protein family A (Hsp70) member 5 |
| chr2_-_89268506 | 1.54 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chr6_-_24489565 | 1.54 |
ENST00000230036.2
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr3_+_184380207 | 1.54 |
ENST00000450923.5
ENST00000348986.3 |
CHRD
|
chordin |
| chr14_-_105626066 | 1.53 |
ENST00000641978.1
ENST00000390543.3 |
IGHG4
|
immunoglobulin heavy constant gamma 4 (G4m marker) |
| chr2_+_90220727 | 1.52 |
ENST00000471857.2
|
IGKV1D-8
|
immunoglobulin kappa variable 1D-8 |
| chr2_-_150487658 | 1.51 |
ENST00000375734.6
ENST00000263895.9 ENST00000454202.5 |
RND3
|
Rho family GTPase 3 |
| chr9_+_27109200 | 1.49 |
ENST00000380036.10
|
TEK
|
TEK receptor tyrosine kinase |
| chr2_+_89862438 | 1.49 |
ENST00000448155.2
|
IGKV1D-39
|
immunoglobulin kappa variable 1D-39 |
| chr3_+_184380047 | 1.44 |
ENST00000204604.5
|
CHRD
|
chordin |
| chrX_+_124346544 | 1.44 |
ENST00000371139.9
|
SH2D1A
|
SH2 domain containing 1A |
| chr22_+_22327298 | 1.43 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr11_-_57390636 | 1.42 |
ENST00000525955.1
ENST00000533605.5 ENST00000311862.10 |
PRG2
|
proteoglycan 2, pro eosinophil major basic protein |
| chr17_-_36001549 | 1.41 |
ENST00000617897.2
|
CCL15
|
C-C motif chemokine ligand 15 |
| chr17_-_75878542 | 1.41 |
ENST00000254816.6
|
TRIM47
|
tripartite motif containing 47 |
| chr22_+_22668286 | 1.40 |
ENST00000390304.2
|
IGLV3-27
|
immunoglobulin lambda variable 3-27 |
| chr17_+_14301069 | 1.38 |
ENST00000360954.3
|
HS3ST3B1
|
heparan sulfate-glucosamine 3-sulfotransferase 3B1 |
| chr4_+_39406917 | 1.38 |
ENST00000257408.5
|
KLB
|
klotho beta |
| chr11_-_5243644 | 1.35 |
ENST00000643122.1
|
HBD
|
hemoglobin subunit delta |
| chr7_+_116672187 | 1.35 |
ENST00000318493.11
ENST00000397752.8 |
MET
|
MET proto-oncogene, receptor tyrosine kinase |
| chr5_+_151020438 | 1.35 |
ENST00000622181.4
ENST00000614343.4 ENST00000388825.9 ENST00000521650.5 ENST00000517973.1 |
GPX3
|
glutathione peroxidase 3 |
| chr6_-_3751703 | 1.34 |
ENST00000380283.5
|
PXDC1
|
PX domain containing 1 |
| chr17_+_1771688 | 1.33 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin family F member 1 |
| chr1_-_201399302 | 1.33 |
ENST00000633953.1
ENST00000391967.7 |
LAD1
|
ladinin 1 |
| chr2_+_20667136 | 1.33 |
ENST00000272224.5
|
GDF7
|
growth differentiation factor 7 |
| chr9_+_27109135 | 1.33 |
ENST00000519097.5
ENST00000615002.4 |
TEK
|
TEK receptor tyrosine kinase |
| chr20_+_35615812 | 1.32 |
ENST00000679710.1
ENST00000374273.8 |
SPAG4
|
sperm associated antigen 4 |
| chr22_+_22322452 | 1.31 |
ENST00000390290.3
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr11_+_70085413 | 1.31 |
ENST00000316296.9
ENST00000530676.5 |
ANO1
|
anoctamin 1 |
| chr14_-_106675544 | 1.31 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr10_+_94762673 | 1.27 |
ENST00000480405.2
ENST00000371321.9 |
CYP2C19
|
cytochrome P450 family 2 subfamily C member 19 |
| chr12_-_14843517 | 1.27 |
ENST00000228936.6
|
ART4
|
ADP-ribosyltransferase 4 (inactive) (Dombrock blood group) |
| chr12_-_102480604 | 1.27 |
ENST00000392905.7
|
IGF1
|
insulin like growth factor 1 |
| chr3_+_50155024 | 1.26 |
ENST00000414301.5
ENST00000450338.5 ENST00000413852.5 |
SEMA3F
|
semaphorin 3F |
| chr1_+_171314171 | 1.26 |
ENST00000367749.4
|
FMO4
|
flavin containing dimethylaniline monoxygenase 4 |
| chr3_-_120682215 | 1.24 |
ENST00000283871.10
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr12_+_52233235 | 1.24 |
ENST00000331817.6
|
KRT7
|
keratin 7 |
| chr17_+_42851167 | 1.24 |
ENST00000613571.1
ENST00000308423.7 |
AOC3
|
amine oxidase copper containing 3 |
| chr12_+_56996151 | 1.23 |
ENST00000556850.1
|
GPR182
|
G protein-coupled receptor 182 |
| chr11_-_914774 | 1.23 |
ENST00000528154.1
ENST00000525840.1 |
CHID1
|
chitinase domain containing 1 |
| chr1_+_61404076 | 1.22 |
ENST00000357977.5
|
NFIA
|
nuclear factor I A |
| chr19_-_7747511 | 1.22 |
ENST00000593660.5
ENST00000204801.12 ENST00000315591.12 ENST00000354397.10 ENST00000394161.9 ENST00000593821.5 ENST00000602261.5 ENST00000601256.1 ENST00000601951.5 ENST00000315599.12 |
CD209
|
CD209 molecule |
| chrX_+_17737443 | 1.20 |
ENST00000398080.5
ENST00000380045.7 ENST00000380043.7 ENST00000380041.8 |
SCML1
|
Scm polycomb group protein like 1 |
| chr11_-_57237183 | 1.20 |
ENST00000606794.1
|
APLNR
|
apelin receptor |
| chr20_+_46709666 | 1.19 |
ENST00000486000.2
|
SLC2A10
|
solute carrier family 2 member 10 |
| chr19_-_3600581 | 1.17 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor |
| chr11_-_2139382 | 1.17 |
ENST00000416167.7
|
IGF2
|
insulin like growth factor 2 |
| chr4_-_57110373 | 1.13 |
ENST00000295666.6
ENST00000514062.2 |
IGFBP7
|
insulin like growth factor binding protein 7 |
| chr3_+_188947199 | 1.12 |
ENST00000433971.5
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr3_+_100609594 | 1.11 |
ENST00000273352.8
|
ADGRG7
|
adhesion G protein-coupled receptor G7 |
| chr2_-_74529670 | 1.10 |
ENST00000377526.4
|
AUP1
|
AUP1 lipid droplet regulating VLDL assembly factor |
| chr22_+_22343185 | 1.10 |
ENST00000427632.2
|
IGLV9-49
|
immunoglobulin lambda variable 9-49 |
| chr2_+_90234809 | 1.10 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr3_+_38346760 | 1.09 |
ENST00000427323.5
ENST00000207870.8 ENST00000650590.1 |
XYLB
|
xylulokinase |
| chr3_+_186567403 | 1.06 |
ENST00000439351.5
|
DNAJB11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr10_-_96271508 | 1.06 |
ENST00000427367.6
ENST00000413476.6 ENST00000371176.6 |
BLNK
|
B cell linker |
| chr11_+_63369779 | 1.05 |
ENST00000279178.4
|
SLC22A9
|
solute carrier family 22 member 9 |
| chr22_+_22375984 | 1.05 |
ENST00000390296.2
|
IGLV5-45
|
immunoglobulin lambda variable 5-45 |
| chr3_+_185363129 | 1.04 |
ENST00000265026.8
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr2_+_27496830 | 1.04 |
ENST00000264717.7
|
GCKR
|
glucokinase regulator |
| chr7_-_41703062 | 1.03 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr6_-_25930678 | 1.03 |
ENST00000377850.8
|
SLC17A2
|
solute carrier family 17 member 2 |
| chr17_+_1762052 | 1.02 |
ENST00000254722.9
ENST00000576406.5 ENST00000571149.5 |
SERPINF1
|
serpin family F member 1 |
| chr15_-_39920162 | 1.02 |
ENST00000299092.3
|
GPR176
|
G protein-coupled receptor 176 |
| chr3_-_150546403 | 1.02 |
ENST00000239944.7
ENST00000491660.1 ENST00000487153.1 |
SERP1
|
stress associated endoplasmic reticulum protein 1 |
| chr21_-_38661694 | 1.01 |
ENST00000417133.6
ENST00000398910.5 ENST00000442448.5 ENST00000429727.6 |
ERG
|
ETS transcription factor ERG |
| chr15_+_80152978 | 1.01 |
ENST00000561421.6
ENST00000684363.1 |
FAH
|
fumarylacetoacetate hydrolase |
| chr2_-_136116165 | 1.01 |
ENST00000409817.1
|
CXCR4
|
C-X-C motif chemokine receptor 4 |
| chr10_-_11611754 | 1.01 |
ENST00000609104.5
|
USP6NL
|
USP6 N-terminal like |
| chr6_-_25930611 | 0.99 |
ENST00000360488.7
|
SLC17A2
|
solute carrier family 17 member 2 |
| chr12_+_55681647 | 0.99 |
ENST00000614691.1
|
METTL7B
|
methyltransferase like 7B |
| chr17_+_76540035 | 0.98 |
ENST00000592014.6
|
PRCD
|
photoreceptor disc component |
| chr12_+_111766887 | 0.97 |
ENST00000416293.7
ENST00000261733.7 |
ALDH2
|
aldehyde dehydrogenase 2 family member |
| chr2_+_236569817 | 0.96 |
ENST00000272928.4
|
ACKR3
|
atypical chemokine receptor 3 |
| chr16_+_8712943 | 0.96 |
ENST00000561870.5
ENST00000396600.6 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr13_+_93226787 | 0.96 |
ENST00000377047.9
|
GPC6
|
glypican 6 |
| chr15_+_96332432 | 0.96 |
ENST00000559679.1
ENST00000394171.6 |
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chr10_+_113553039 | 0.95 |
ENST00000351270.4
|
HABP2
|
hyaluronan binding protein 2 |
| chr6_+_33200820 | 0.95 |
ENST00000374675.7
|
SLC39A7
|
solute carrier family 39 member 7 |
| chr2_+_233729042 | 0.95 |
ENST00000482026.6
|
UGT1A3
|
UDP glucuronosyltransferase family 1 member A3 |
| chr2_-_64144411 | 0.94 |
ENST00000358912.5
|
PELI1
|
pellino E3 ubiquitin protein ligase 1 |
| chr1_-_42766978 | 0.94 |
ENST00000372526.2
ENST00000236040.8 ENST00000296388.10 ENST00000397054.7 |
P3H1
|
prolyl 3-hydroxylase 1 |
| chr9_+_136662907 | 0.94 |
ENST00000308874.12
ENST00000406555.7 ENST00000492862.6 |
EGFL7
|
EGF like domain multiple 7 |
| chr12_+_55681711 | 0.94 |
ENST00000394252.4
|
METTL7B
|
methyltransferase like 7B |
| chr9_-_109498251 | 0.92 |
ENST00000374541.4
ENST00000262539.7 |
PTPN3
|
protein tyrosine phosphatase non-receptor type 3 |
| chr3_-_64687992 | 0.90 |
ENST00000498707.5
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif 9 |
| chr22_+_22162155 | 0.89 |
ENST00000390284.2
|
IGLV4-60
|
immunoglobulin lambda variable 4-60 |
| chr11_-_124762283 | 0.89 |
ENST00000444566.5
ENST00000278927.10 ENST00000435477.1 |
ESAM
|
endothelial cell adhesion molecule |
| chr7_+_40134966 | 0.89 |
ENST00000401647.7
ENST00000628514.3 ENST00000335693.9 ENST00000416370.2 |
SUGCT
|
succinyl-CoA:glutarate-CoA transferase |
| chr3_+_50155305 | 0.88 |
ENST00000002829.8
ENST00000426511.5 |
SEMA3F
|
semaphorin 3F |
| chr6_-_33329262 | 0.87 |
ENST00000494082.3
|
SMIM40
|
small integral membrane protein 40 |
| chr3_-_123620571 | 0.86 |
ENST00000583087.5
|
MYLK
|
myosin light chain kinase |
| chr1_-_12616762 | 0.85 |
ENST00000464917.5
|
DHRS3
|
dehydrogenase/reductase 3 |
| chr11_-_118252279 | 0.84 |
ENST00000525386.5
ENST00000527472.1 ENST00000278949.9 |
MPZL3
|
myelin protein zero like 3 |
| chr12_-_11269696 | 0.83 |
ENST00000381842.7
|
PRB3
|
proline rich protein BstNI subfamily 3 |
| chr1_-_8879170 | 0.83 |
ENST00000489867.2
|
ENO1
|
enolase 1 |
| chr8_+_30387064 | 0.83 |
ENST00000523115.5
ENST00000519647.5 |
RBPMS
|
RNA binding protein, mRNA processing factor |
| chr10_+_119207560 | 0.83 |
ENST00000392870.3
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr19_-_54257217 | 0.82 |
ENST00000345866.10
ENST00000449561.3 |
LILRB5
|
leukocyte immunoglobulin like receptor B5 |
| chr19_-_7747559 | 0.81 |
ENST00000394173.8
|
CD209
|
CD209 molecule |
| chrX_+_129779930 | 0.81 |
ENST00000356892.4
|
SASH3
|
SAM and SH3 domain containing 3 |
| chr1_+_212285383 | 0.81 |
ENST00000261461.7
|
PPP2R5A
|
protein phosphatase 2 regulatory subunit B'alpha |
| chr16_+_11965234 | 0.80 |
ENST00000562385.1
|
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr19_+_35115912 | 0.80 |
ENST00000603181.5
|
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr20_-_62367304 | 0.80 |
ENST00000252999.7
|
LAMA5
|
laminin subunit alpha 5 |
| chrX_+_124346525 | 0.79 |
ENST00000360027.4
|
SH2D1A
|
SH2 domain containing 1A |
| chr7_+_26291850 | 0.79 |
ENST00000338523.9
ENST00000446848.6 |
SNX10
|
sorting nexin 10 |
| chr3_-_64687982 | 0.78 |
ENST00000459780.1
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif 9 |
| chr7_-_120857124 | 0.78 |
ENST00000441017.5
ENST00000424710.5 ENST00000433758.5 |
TSPAN12
|
tetraspanin 12 |
| chr20_+_43558968 | 0.78 |
ENST00000647834.1
ENST00000373100.7 ENST00000648083.1 ENST00000648530.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr1_-_109392893 | 0.78 |
ENST00000633956.1
|
SORT1
|
sortilin 1 |
| chr6_+_33200860 | 0.78 |
ENST00000374677.8
|
SLC39A7
|
solute carrier family 39 member 7 |
| chr15_-_39920248 | 0.78 |
ENST00000561100.2
|
GPR176
|
G protein-coupled receptor 176 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAF_NRL
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 19.8 | GO:2000910 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 3.6 | 14.5 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 3.6 | 3.6 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 2.7 | 10.6 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 1.4 | 25.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.3 | 12.1 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 1.2 | 7.5 | GO:0008218 | bioluminescence(GO:0008218) |
| 1.0 | 5.2 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.7 | 6.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.7 | 2.0 | GO:1902688 | regulation of NAD metabolic process(GO:1902688) |
| 0.6 | 6.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.6 | 1.8 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.6 | 8.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.5 | 2.0 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.5 | 2.0 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.5 | 5.0 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.5 | 2.7 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.4 | 5.5 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.4 | 2.5 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.4 | 2.5 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.4 | 1.2 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.4 | 1.5 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.4 | 3.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.4 | 1.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.4 | 1.4 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.4 | 8.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.4 | 1.8 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 1.0 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.3 | 1.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.3 | 2.0 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 0.9 | GO:1901874 | negative regulation of post-translational protein modification(GO:1901874) |
| 0.3 | 1.6 | GO:1903891 | regulation of ATF6-mediated unfolded protein response(GO:1903891) |
| 0.3 | 2.4 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.3 | 41.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.3 | 0.8 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.3 | 2.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.3 | 0.8 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.2 | 0.7 | GO:0046247 | tetraterpenoid metabolic process(GO:0016108) carotenoid metabolic process(GO:0016116) carotene catabolic process(GO:0016121) xanthophyll metabolic process(GO:0016122) terpene catabolic process(GO:0046247) |
| 0.2 | 1.0 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.2 | 1.0 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 0.5 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.2 | 1.6 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.2 | 0.7 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.2 | 1.8 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 | 1.6 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) |
| 0.2 | 0.9 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 0.6 | GO:0071650 | negative regulation of interleukin-18 production(GO:0032701) negative regulation of cytokine activity(GO:0060302) negative regulation of chemokine (C-C motif) ligand 5 production(GO:0071650) |
| 0.2 | 0.9 | GO:0035548 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) negative regulation of CD8-positive, alpha-beta T cell activation(GO:2001186) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.2 | 3.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 1.2 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.2 | 1.6 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 1.4 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.2 | 0.8 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.2 | 2.1 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.2 | 1.3 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.2 | 0.8 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.6 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 0.7 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.2 | 0.5 | GO:0036323 | vascular endothelial growth factor receptor-1 signaling pathway(GO:0036323) |
| 0.2 | 1.6 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 0.7 | GO:0010193 | response to ozone(GO:0010193) |
| 0.2 | 1.9 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.2 | 2.0 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.2 | 1.0 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.2 | 2.9 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.2 | 1.3 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 11.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 1.3 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.2 | 1.5 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.2 | 0.3 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.2 | 0.9 | GO:0001554 | luteolysis(GO:0001554) |
| 0.2 | 0.5 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.1 | 1.0 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 4.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 1.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 1.7 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.7 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.4 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.1 | 1.7 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.5 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.4 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.1 | 2.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.5 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 0.4 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 1.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.6 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 | 0.3 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 1.3 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.1 | 0.6 | GO:0042851 | L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.1 | 0.7 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 0.7 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 2.7 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.6 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 0.7 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.9 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.8 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 0.4 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.3 | GO:1903414 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.1 | 0.4 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.1 | 0.6 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.4 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.1 | 0.4 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 0.8 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.3 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.3 | GO:0036047 | protein demalonylation(GO:0036046) peptidyl-lysine demalonylation(GO:0036047) protein desuccinylation(GO:0036048) peptidyl-lysine desuccinylation(GO:0036049) protein deglutarylation(GO:0061698) peptidyl-lysine deglutarylation(GO:0061699) |
| 0.1 | 0.7 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.8 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.1 | 0.4 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 4.0 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 2.9 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.6 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 | 0.2 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.1 | 0.3 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 0.6 | GO:2000230 | regulation of cholesterol transporter activity(GO:0060694) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 1.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.2 | GO:0060829 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 1.3 | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.1 | 2.4 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.1 | 0.4 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.1 | 0.6 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.9 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.6 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.1 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.1 | 0.3 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 2.0 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.2 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 3.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.4 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 3.7 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 0.7 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.1 | 0.4 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.1 | 0.7 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.1 | 0.9 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.1 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.3 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.1 | 0.8 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 12.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.2 | GO:1903984 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 1.9 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.6 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 0.4 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.2 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.5 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.3 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 1.2 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.1 | 0.6 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.1 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.0 | 1.0 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.8 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.5 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.0 | 0.7 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.4 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 4.5 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.4 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.0 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.6 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.1 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.0 | 0.2 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 0.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.0 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 1.3 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.0 | 0.4 | GO:0002254 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.3 | GO:0046102 | inosine metabolic process(GO:0046102) |
| 0.0 | 2.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.8 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 2.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.5 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.6 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.4 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.5 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.3 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.4 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.3 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.6 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 1.1 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.5 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.6 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.4 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 2.2 | GO:0030818 | negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.0 | 0.1 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.2 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.5 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.6 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.7 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 1.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 1.0 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.5 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 2.0 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.4 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.4 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.3 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 1.2 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.0 | 0.6 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.2 | GO:2000018 | regulation of male gonad development(GO:2000018) positive regulation of male gonad development(GO:2000020) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 0.3 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.3 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.2 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.0 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.3 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.1 | GO:0070861 | regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 2.1 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0001971 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.0 | 0.3 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 1.2 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.8 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.5 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 1.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.6 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 1.0 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.9 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 23.5 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 2.8 | 25.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.0 | 4.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.9 | 2.7 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.7 | 14.9 | GO:0042627 | chylomicron(GO:0042627) |
| 0.4 | 4.4 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.3 | 1.0 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 16.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 0.8 | GO:0043259 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.3 | 0.8 | GO:1990427 | stereocilia tip link(GO:0002140) myosin VII complex(GO:0031477) stereocilia tip-link density(GO:1990427) upper tip-link density(GO:1990435) |
| 0.3 | 5.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.2 | 1.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 1.4 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.2 | 0.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 31.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 0.8 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.6 | GO:0070701 | mucus layer(GO:0070701) |
| 0.1 | 0.7 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.4 | GO:0060342 | photoreceptor inner segment membrane(GO:0060342) |
| 0.1 | 8.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.4 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 0.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 1.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.4 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.2 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.7 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.5 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 0.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.6 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 1.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 2.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.0 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.7 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.5 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.8 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 18.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.4 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 2.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 61.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 2.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.4 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 3.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 1.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.9 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.3 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 23.5 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 2.1 | 10.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.8 | 14.5 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.8 | 5.5 | GO:0019862 | IgA binding(GO:0019862) |
| 0.7 | 2.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.6 | 1.9 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.5 | 2.7 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.4 | 8.9 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.4 | 1.2 | GO:0004411 | homogentisate 1,2-dioxygenase activity(GO:0004411) |
| 0.4 | 1.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.4 | 1.2 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.4 | 2.5 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.3 | 2.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.3 | 3.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 1.6 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.3 | 0.6 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.3 | 1.1 | GO:0042282 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.3 | 0.8 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.2 | 1.6 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 0.7 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.2 | 2.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 1.3 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 0.9 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.2 | 1.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 13.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 1.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 1.4 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.2 | 0.8 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 0.6 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.2 | 1.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 0.9 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.2 | 0.6 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.2 | 0.6 | GO:0051990 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.2 | 4.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 1.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.5 | GO:0036332 | VEGF-A-activated receptor activity(GO:0036326) VEGF-B-activated receptor activity(GO:0036327) placental growth factor-activated receptor activity(GO:0036332) |
| 0.2 | 6.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 28.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 1.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 0.7 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 1.5 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.2 | 3.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 0.8 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 0.6 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.2 | 0.8 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.2 | 0.5 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.1 | 1.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 3.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 31.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.8 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.8 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 2.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.4 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.1 | 0.4 | GO:0004577 | N-acetylglucosaminyldiphosphodolichol N-acetylglucosaminyltransferase activity(GO:0004577) |
| 0.1 | 0.4 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 3.7 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 2.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.3 | GO:0019808 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.1 | 0.8 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 1.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.7 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 5.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.5 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.8 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.3 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 1.6 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 2.0 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.3 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.1 | 0.3 | GO:0036054 | protein-malonyllysine demalonylase activity(GO:0036054) protein-succinyllysine desuccinylase activity(GO:0036055) protein-glutaryllysine deglutarylase activity(GO:0061697) |
| 0.1 | 5.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.5 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.6 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 1.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 1.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.2 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.1 | 0.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.4 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 2.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.2 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 1.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 1.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.2 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.1 | 1.0 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.3 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 5.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.5 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.3 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.1 | 0.5 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.4 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.1 | 0.3 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.5 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 1.0 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.6 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 1.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 2.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.4 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 6.8 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 0.7 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 1.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 13.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 3.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.6 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 2.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.9 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.3 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 1.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 1.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.7 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 3.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 1.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 2.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.0 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 1.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 2.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.0 | GO:0008083 | growth factor activity(GO:0008083) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 12.9 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 10.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 8.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 9.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 34.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 6.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 7.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 0.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 2.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 4.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 3.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 2.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 2.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.9 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.5 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.9 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 1.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 3.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 23.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.9 | 44.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.5 | 8.9 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.3 | 7.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 5.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 8.5 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.2 | 1.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 2.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 4.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 9.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 4.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 2.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 2.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 0.9 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.0 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.7 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 2.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 2.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 2.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 2.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.9 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 6.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.9 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.5 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |