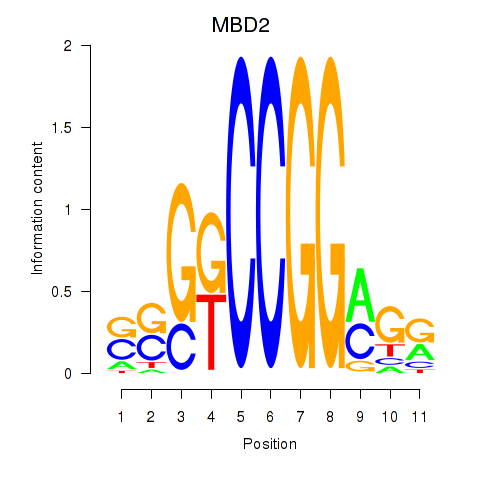
|
chr10_-_17617326
Show fit
|
1.43 |
ENST00000326961.6
ENST00000361271.8
|
HACD1
|
3-hydroxyacyl-CoA dehydratase 1
|
|
chr4_-_6472548
Show fit
|
1.17 |
ENST00000382599.9
|
PPP2R2C
|
protein phosphatase 2 regulatory subunit Bgamma
|
|
chr2_+_219434825
Show fit
|
1.17 |
ENST00000312358.12
|
SPEG
|
striated muscle enriched protein kinase
|
|
chr1_+_109466527
Show fit
|
1.17 |
ENST00000369872.4
|
SYPL2
|
synaptophysin like 2
|
|
chr9_-_109498251
Show fit
|
1.12 |
ENST00000374541.4
ENST00000262539.7
|
PTPN3
|
protein tyrosine phosphatase non-receptor type 3
|
|
chr18_-_28177934
Show fit
|
1.12 |
ENST00000676445.1
|
CDH2
|
cadherin 2
|
|
chrX_+_69504320
Show fit
|
1.10 |
ENST00000252338.5
|
FAM155B
|
family with sequence similarity 155 member B
|
|
chr10_-_17617235
Show fit
|
1.07 |
ENST00000466335.1
|
HACD1
|
3-hydroxyacyl-CoA dehydratase 1
|
|
chr11_-_46918522
Show fit
|
1.03 |
ENST00000378623.6
ENST00000534404.1
|
LRP4
|
LDL receptor related protein 4
|
|
chr17_-_39197696
Show fit
|
1.02 |
ENST00000394310.7
ENST00000622445.4
ENST00000344140.5
|
CACNB1
|
calcium voltage-gated channel auxiliary subunit beta 1
|
|
chr7_-_51316754
Show fit
|
1.01 |
ENST00000632460.1
ENST00000441453.5
ENST00000648294.1
ENST00000265136.12
ENST00000395542.6
ENST00000395540.6
|
COBL
|
cordon-bleu WH2 repeat protein
|
|
chr3_-_142963663
Show fit
|
0.99 |
ENST00000340634.6
|
PAQR9
|
progestin and adipoQ receptor family member 9
|
|
chr16_-_51151259
Show fit
|
0.98 |
ENST00000251020.9
|
SALL1
|
spalt like transcription factor 1
|
|
chr17_-_29622893
Show fit
|
0.91 |
ENST00000345068.9
ENST00000584602.1
ENST00000388767.8
ENST00000580212.6
|
CORO6
|
coronin 6
|
|
chr19_+_55283982
Show fit
|
0.84 |
ENST00000309383.6
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr7_+_141074038
Show fit
|
0.83 |
ENST00000565468.6
ENST00000610315.1
|
TMEM178B
|
transmembrane protein 178B
|
|
chr18_+_50560070
Show fit
|
0.81 |
ENST00000400384.7
ENST00000540640.3
ENST00000592595.5
|
MAPK4
|
mitogen-activated protein kinase 4
|
|
chr7_-_100895878
Show fit
|
0.81 |
ENST00000419336.6
ENST00000241069.11
ENST00000411582.4
ENST00000302913.8
|
ACHE
|
acetylcholinesterase (Cartwright blood group)
|
|
chr15_-_65422894
Show fit
|
0.78 |
ENST00000352385.3
|
IGDCC4
|
immunoglobulin superfamily DCC subclass member 4
|
|
chr10_+_115093331
Show fit
|
0.73 |
ENST00000609571.5
ENST00000355044.8
ENST00000526946.5
|
ATRNL1
|
attractin like 1
|
|
chr17_-_39197652
Show fit
|
0.72 |
ENST00000394303.8
|
CACNB1
|
calcium voltage-gated channel auxiliary subunit beta 1
|
|
chr17_-_29622732
Show fit
|
0.72 |
ENST00000683819.1
ENST00000492276.7
|
CORO6
|
coronin 6
|
|
chr16_-_51151238
Show fit
|
0.72 |
ENST00000566102.1
|
SALL1
|
spalt like transcription factor 1
|
|
chr7_-_150978284
Show fit
|
0.72 |
ENST00000262186.10
|
KCNH2
|
potassium voltage-gated channel subfamily H member 2
|
|
chr15_-_73753308
Show fit
|
0.70 |
ENST00000569673.3
|
INSYN1
|
inhibitory synaptic factor 1
|
|
chr15_-_101489697
Show fit
|
0.70 |
ENST00000611967.4
ENST00000615296.4
ENST00000611716.5
ENST00000618548.4
ENST00000619160.4
ENST00000622483.4
ENST00000559417.2
|
PCSK6
|
proprotein convertase subtilisin/kexin type 6
|
|
chr4_-_52659238
Show fit
|
0.67 |
ENST00000451218.6
ENST00000441222.8
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr16_-_81096163
Show fit
|
0.64 |
ENST00000566566.2
ENST00000569885.6
ENST00000561801.2
ENST00000639689.1
ENST00000638948.1
ENST00000564536.2
ENST00000638192.1
ENST00000640345.1
|
GCSH
ENSG00000260643.2
ENSG00000284512.1
|
glycine cleavage system protein H
novel protein
novel protein
|
|
chr1_-_231422261
Show fit
|
0.62 |
ENST00000366641.4
|
EGLN1
|
egl-9 family hypoxia inducible factor 1
|
|
chr7_-_100896123
Show fit
|
0.62 |
ENST00000428317.7
|
ACHE
|
acetylcholinesterase (Cartwright blood group)
|
|
chr2_+_100820102
Show fit
|
0.61 |
ENST00000335681.10
|
NPAS2
|
neuronal PAS domain protein 2
|
|
chr2_+_197804583
Show fit
|
0.61 |
ENST00000428675.6
|
PLCL1
|
phospholipase C like 1 (inactive)
|
|
chr18_+_8609404
Show fit
|
0.60 |
ENST00000649141.2
|
RAB12
|
RAB12, member RAS oncogene family
|
|
chr7_+_74453790
Show fit
|
0.60 |
ENST00000265755.7
ENST00000424337.7
ENST00000455841.6
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr9_+_34989641
Show fit
|
0.59 |
ENST00000453597.8
ENST00000312316.9
ENST00000458263.6
ENST00000537321.5
ENST00000682809.1
ENST00000684748.1
|
DNAJB5
|
DnaJ heat shock protein family (Hsp40) member B5
|
|
chr17_-_6556447
Show fit
|
0.59 |
ENST00000421306.7
|
PITPNM3
|
PITPNM family member 3
|
|
chr1_-_31763866
Show fit
|
0.58 |
ENST00000398547.5
|
ADGRB2
|
adhesion G protein-coupled receptor B2
|
|
chr17_+_50561010
Show fit
|
0.58 |
ENST00000360761.8
ENST00000354983.8
ENST00000352832.9
|
CACNA1G
|
calcium voltage-gated channel subunit alpha1 G
|
|
chr15_-_73633234
Show fit
|
0.57 |
ENST00000562924.5
ENST00000563691.5
ENST00000565325.5
|
NPTN
|
neuroplastin
|
|
chr11_+_66258467
Show fit
|
0.57 |
ENST00000394066.6
|
KLC2
|
kinesin light chain 2
|
|
chr1_+_52142044
Show fit
|
0.56 |
ENST00000287727.8
ENST00000371591.2
|
ZFYVE9
|
zinc finger FYVE-type containing 9
|
|
chr1_-_31764333
Show fit
|
0.55 |
ENST00000398542.5
ENST00000373658.8
|
ADGRB2
|
adhesion G protein-coupled receptor B2
|
|
chr6_+_125154189
Show fit
|
0.55 |
ENST00000532429.5
ENST00000534199.5
|
TPD52L1
|
TPD52 like 1
|
|
chr13_-_75482114
Show fit
|
0.54 |
ENST00000377625.6
ENST00000431480.6
|
TBC1D4
|
TBC1 domain family member 4
|
|
chr11_-_45665578
Show fit
|
0.53 |
ENST00000308064.7
|
CHST1
|
carbohydrate sulfotransferase 1
|
|
chr1_-_22143088
Show fit
|
0.53 |
ENST00000290167.11
|
WNT4
|
Wnt family member 4
|
|
chr22_+_19714675
Show fit
|
0.53 |
ENST00000412544.5
|
SEPTIN5
|
septin 5
|
|
chr1_-_31764035
Show fit
|
0.53 |
ENST00000373655.6
|
ADGRB2
|
adhesion G protein-coupled receptor B2
|
|
chr13_-_75482151
Show fit
|
0.52 |
ENST00000377636.8
|
TBC1D4
|
TBC1 domain family member 4
|
|
chr14_+_52553273
Show fit
|
0.52 |
ENST00000542169.6
ENST00000555622.1
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr6_-_46015607
Show fit
|
0.52 |
ENST00000644878.1
ENST00000644324.1
ENST00000672327.1
|
CLIC5
|
chloride intracellular channel 5
|
|
chr22_+_28883564
Show fit
|
0.52 |
ENST00000544604.7
|
ZNRF3
|
zinc and ring finger 3
|
|
chrX_+_153687918
Show fit
|
0.51 |
ENST00000253122.10
|
SLC6A8
|
solute carrier family 6 member 8
|
|
chr6_+_89433164
Show fit
|
0.51 |
ENST00000369408.9
ENST00000447838.6
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr1_+_6785437
Show fit
|
0.50 |
ENST00000303635.12
ENST00000473578.5
ENST00000557126.5
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr3_-_51967410
Show fit
|
0.50 |
ENST00000461554.6
ENST00000483411.5
ENST00000461544.2
ENST00000355852.6
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr6_+_89433127
Show fit
|
0.50 |
ENST00000339746.9
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr22_-_19431692
Show fit
|
0.50 |
ENST00000340170.8
ENST00000263208.5
|
HIRA
|
histone cell cycle regulator
|
|
chr2_+_42048012
Show fit
|
0.50 |
ENST00000294964.6
|
PKDCC
|
protein kinase domain containing, cytoplasmic
|
|
chr9_+_100427254
Show fit
|
0.49 |
ENST00000374885.5
|
MSANTD3
|
Myb/SANT DNA binding domain containing 3
|
|
chr2_+_238426920
Show fit
|
0.49 |
ENST00000264607.9
|
ASB1
|
ankyrin repeat and SOCS box containing 1
|
|
chr10_-_124744280
Show fit
|
0.49 |
ENST00000337318.8
|
FAM53B
|
family with sequence similarity 53 member B
|
|
chr22_+_19714450
Show fit
|
0.48 |
ENST00000455784.7
ENST00000406395.5
|
SEPTIN5
|
septin 5
|
|
chr20_-_3159844
Show fit
|
0.47 |
ENST00000217173.7
ENST00000449731.1
ENST00000380266.4
|
UBOX5
FASTKD5
|
U-box domain containing 5
FAST kinase domains 5
|
|
chr6_+_163414701
Show fit
|
0.47 |
ENST00000361752.8
|
QKI
|
QKI, KH domain containing RNA binding
|
|
chr11_+_45146631
Show fit
|
0.45 |
ENST00000534751.3
ENST00000683152.1
|
PRDM11
|
PR/SET domain 11
|
|
chr17_+_50560703
Show fit
|
0.44 |
ENST00000359106.10
|
CACNA1G
|
calcium voltage-gated channel subunit alpha1 G
|
|
chr14_-_77320741
Show fit
|
0.44 |
ENST00000682795.1
ENST00000682247.1
|
POMT2
|
protein O-mannosyltransferase 2
|
|
chr2_-_118014125
Show fit
|
0.44 |
ENST00000319432.9
|
CCDC93
|
coiled-coil domain containing 93
|
|
chr14_-_23578756
Show fit
|
0.43 |
ENST00000397118.7
ENST00000356300.9
|
JPH4
|
junctophilin 4
|
|
chr11_-_34357994
Show fit
|
0.43 |
ENST00000435224.3
|
ABTB2
|
ankyrin repeat and BTB domain containing 2
|
|
chr20_-_44810539
Show fit
|
0.43 |
ENST00000372851.8
|
RIMS4
|
regulating synaptic membrane exocytosis 4
|
|
chr1_-_247331743
Show fit
|
0.42 |
ENST00000294753.8
ENST00000682384.1
|
ZNF496
|
zinc finger protein 496
|
|
chr19_+_36054879
Show fit
|
0.42 |
ENST00000378860.8
ENST00000427823.3
ENST00000681542.1
ENST00000680321.1
ENST00000680359.1
ENST00000680403.1
ENST00000679714.1
ENST00000679682.1
ENST00000401500.7
ENST00000680564.1
|
WDR62
|
WD repeat domain 62
|
|
chr16_+_70114306
Show fit
|
0.41 |
ENST00000288050.9
ENST00000398122.7
ENST00000568530.5
|
PDPR
|
pyruvate dehydrogenase phosphatase regulatory subunit
|
|
chr20_+_37903104
Show fit
|
0.41 |
ENST00000373459.4
ENST00000373461.9
ENST00000448944.1
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chr12_+_125186376
Show fit
|
0.40 |
ENST00000682704.1
|
TMEM132B
|
transmembrane protein 132B
|
|
chr2_-_118014035
Show fit
|
0.40 |
ENST00000376300.7
|
CCDC93
|
coiled-coil domain containing 93
|
|
chr3_+_54122542
Show fit
|
0.39 |
ENST00000415676.6
ENST00000474759.6
|
CACNA2D3
|
calcium voltage-gated channel auxiliary subunit alpha2delta 3
|
|
chr8_-_73747458
Show fit
|
0.39 |
ENST00000518767.5
|
STAU2
|
staufen double-stranded RNA binding protein 2
|
|
chr19_+_3359563
Show fit
|
0.39 |
ENST00000589123.5
ENST00000395111.7
ENST00000586919.5
|
NFIC
|
nuclear factor I C
|
|
chr11_-_18791563
Show fit
|
0.39 |
ENST00000396168.1
|
PTPN5
|
protein tyrosine phosphatase non-receptor type 5
|
|
chr6_-_46015812
Show fit
|
0.39 |
ENST00000544153.3
ENST00000339561.12
|
CLIC5
|
chloride intracellular channel 5
|
|
chr11_-_110296627
Show fit
|
0.39 |
ENST00000533991.1
|
RDX
|
radixin
|
|
chr15_-_42491105
Show fit
|
0.38 |
ENST00000565380.5
ENST00000564754.7
|
ZNF106
|
zinc finger protein 106
|
|
chr12_+_19129689
Show fit
|
0.38 |
ENST00000429027.7
ENST00000540972.5
|
PLEKHA5
|
pleckstrin homology domain containing A5
|
|
chr11_-_27700472
Show fit
|
0.38 |
ENST00000418212.5
ENST00000533246.5
|
BDNF
|
brain derived neurotrophic factor
|
|
chr6_-_166308385
Show fit
|
0.38 |
ENST00000322583.5
|
PRR18
|
proline rich 18
|
|
chr1_+_6785518
Show fit
|
0.38 |
ENST00000467404.6
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr5_+_176810498
Show fit
|
0.37 |
ENST00000509580.2
|
UNC5A
|
unc-5 netrin receptor A
|
|
chr18_-_76990002
Show fit
|
0.37 |
ENST00000585201.5
|
MBP
|
myelin basic protein
|
|
chr9_+_135075520
Show fit
|
0.37 |
ENST00000252854.8
|
OLFM1
|
olfactomedin 1
|
|
chr19_+_3366549
Show fit
|
0.37 |
ENST00000341919.7
ENST00000590282.5
ENST00000443272.3
|
NFIC
|
nuclear factor I C
|
|
chr15_-_73633310
Show fit
|
0.36 |
ENST00000345330.9
|
NPTN
|
neuroplastin
|
|
chr1_+_3624978
Show fit
|
0.36 |
ENST00000378344.7
ENST00000344579.5
|
TPRG1L
|
tumor protein p63 regulated 1 like
|
|
chr17_-_6556539
Show fit
|
0.36 |
ENST00000262483.13
|
PITPNM3
|
PITPNM family member 3
|
|
chr13_+_34942263
Show fit
|
0.35 |
ENST00000379939.7
ENST00000400445.7
|
NBEA
|
neurobeachin
|
|
chr5_+_98773651
Show fit
|
0.35 |
ENST00000513185.3
|
RGMB
|
repulsive guidance molecule BMP co-receptor b
|
|
chr15_+_90001300
Show fit
|
0.35 |
ENST00000268154.9
|
ZNF710
|
zinc finger protein 710
|
|
chr13_+_20703677
Show fit
|
0.34 |
ENST00000682841.1
|
IL17D
|
interleukin 17D
|
|
chr3_-_123449083
Show fit
|
0.34 |
ENST00000462833.6
|
ADCY5
|
adenylate cyclase 5
|
|
chr17_+_76868396
Show fit
|
0.34 |
ENST00000569840.7
|
MGAT5B
|
alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase B
|
|
chr11_-_18791768
Show fit
|
0.33 |
ENST00000358540.7
|
PTPN5
|
protein tyrosine phosphatase non-receptor type 5
|
|
chr1_-_153946652
Show fit
|
0.33 |
ENST00000361217.9
|
DENND4B
|
DENN domain containing 4B
|
|
chr11_-_27700447
Show fit
|
0.33 |
ENST00000356660.9
|
BDNF
|
brain derived neurotrophic factor
|
|
chr2_-_99489955
Show fit
|
0.33 |
ENST00000393445.7
ENST00000258428.8
|
REV1
|
REV1 DNA directed polymerase
|
|
chr19_-_17334804
Show fit
|
0.33 |
ENST00000630631.1
|
ANO8
|
anoctamin 8
|
|
chr11_-_134412234
Show fit
|
0.33 |
ENST00000312527.9
|
B3GAT1
|
beta-1,3-glucuronyltransferase 1
|
|
chrX_-_63755032
Show fit
|
0.33 |
ENST00000624538.2
ENST00000636276.1
ENST00000624843.3
ENST00000671907.1
ENST00000624210.3
ENST00000374870.8
ENST00000635967.1
ENST00000253401.10
ENST00000672194.1
ENST00000637723.2
ENST00000637417.1
ENST00000637520.1
ENST00000374872.4
ENST00000636926.1
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9
|
|
chr17_+_29593468
Show fit
|
0.32 |
ENST00000614878.4
|
ANKRD13B
|
ankyrin repeat domain 13B
|
|
chr1_-_35557378
Show fit
|
0.32 |
ENST00000325722.8
ENST00000469892.5
|
KIAA0319L
|
KIAA0319 like
|
|
chr16_-_28063704
Show fit
|
0.32 |
ENST00000447459.7
|
GSG1L
|
GSG1 like
|
|
chr1_-_53328053
Show fit
|
0.32 |
ENST00000371454.6
ENST00000667377.1
ENST00000306052.12
ENST00000668448.1
|
LRP8
|
LDL receptor related protein 8
|
|
chr19_+_5805018
Show fit
|
0.32 |
ENST00000303212.3
|
NRTN
|
neurturin
|
|
chr22_+_44024269
Show fit
|
0.32 |
ENST00000338758.12
|
PARVB
|
parvin beta
|
|
chr1_-_31239880
Show fit
|
0.32 |
ENST00000373736.7
|
NKAIN1
|
sodium/potassium transporting ATPase interacting 1
|
|
chr19_-_11197516
Show fit
|
0.32 |
ENST00000592903.5
ENST00000586659.6
ENST00000589359.5
ENST00000588724.5
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr9_+_100427176
Show fit
|
0.32 |
ENST00000398977.2
|
MSANTD3
|
Myb/SANT DNA binding domain containing 3
|
|
chr14_+_73950489
Show fit
|
0.32 |
ENST00000554320.1
|
COQ6
|
coenzyme Q6, monooxygenase
|
|
chr17_+_76868410
Show fit
|
0.32 |
ENST00000301618.8
|
MGAT5B
|
alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase B
|
|
chr3_-_197297523
Show fit
|
0.32 |
ENST00000434148.1
ENST00000412364.2
ENST00000661013.1
ENST00000666007.1
ENST00000422288.6
ENST00000456699.6
ENST00000392380.6
ENST00000670935.1
ENST00000656087.1
ENST00000436682.6
ENST00000662727.1
ENST00000670455.1
ENST00000659221.1
ENST00000671185.1
ENST00000669565.1
ENST00000660898.1
ENST00000667971.1
ENST00000661453.1
|
DLG1
|
discs large MAGUK scaffold protein 1
|
|
chr12_+_55743283
Show fit
|
0.32 |
ENST00000546799.1
|
GDF11
|
growth differentiation factor 11
|
|
chr3_+_54122636
Show fit
|
0.32 |
ENST00000288197.9
|
CACNA2D3
|
calcium voltage-gated channel auxiliary subunit alpha2delta 3
|
|
chr22_-_20016807
Show fit
|
0.32 |
ENST00000263207.8
|
ARVCF
|
ARVCF delta catenin family member
|
|
chr12_-_42238261
Show fit
|
0.32 |
ENST00000380790.4
|
YAF2
|
YY1 associated factor 2
|
|
chr2_-_183038405
Show fit
|
0.31 |
ENST00000361354.9
|
NCKAP1
|
NCK associated protein 1
|
|
chr12_-_42238203
Show fit
|
0.31 |
ENST00000327791.8
ENST00000534854.7
|
YAF2
|
YY1 associated factor 2
|
|
chrX_+_153072454
Show fit
|
0.31 |
ENST00000421798.5
|
PNMA6A
|
PNMA family member 6A
|
|
chr1_-_154558650
Show fit
|
0.31 |
ENST00000292211.5
|
UBE2Q1
|
ubiquitin conjugating enzyme E2 Q1
|
|
chr2_+_26970628
Show fit
|
0.31 |
ENST00000233121.7
ENST00000405074.7
|
MAPRE3
|
microtubule associated protein RP/EB family member 3
|
|
chr18_+_74597850
Show fit
|
0.31 |
ENST00000582337.5
ENST00000299687.10
|
ZNF407
|
zinc finger protein 407
|
|
chr7_-_28958321
Show fit
|
0.31 |
ENST00000539664.3
|
TRIL
|
TLR4 interactor with leucine rich repeats
|
|
chr19_+_36054930
Show fit
|
0.31 |
ENST00000679757.1
ENST00000270301.12
|
WDR62
|
WD repeat domain 62
|
|
chr14_-_77320813
Show fit
|
0.30 |
ENST00000682467.1
|
POMT2
|
protein O-mannosyltransferase 2
|
|
chr13_+_21671067
Show fit
|
0.30 |
ENST00000382353.6
|
FGF9
|
fibroblast growth factor 9
|
|
chr10_+_124801799
Show fit
|
0.30 |
ENST00000298492.6
|
ABRAXAS2
|
abraxas 2, BRISC complex subunit
|
|
chr10_-_103855406
Show fit
|
0.30 |
ENST00000355946.6
ENST00000369774.8
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr7_+_43112593
Show fit
|
0.30 |
ENST00000453890.5
ENST00000395891.7
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1
|
|
chr12_+_19129779
Show fit
|
0.30 |
ENST00000539256.5
ENST00000299275.10
ENST00000538714.5
|
PLEKHA5
|
pleckstrin homology domain containing A5
|
|
chr20_+_62302896
Show fit
|
0.30 |
ENST00000620230.4
ENST00000253003.7
|
ADRM1
|
adhesion regulating molecule 1
|
|
chr1_+_179025804
Show fit
|
0.30 |
ENST00000440702.5
|
FAM20B
|
FAM20B glycosaminoglycan xylosylkinase
|
|
chrX_-_63755187
Show fit
|
0.29 |
ENST00000635729.1
ENST00000623566.3
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9
|
|
chr11_-_134411854
Show fit
|
0.29 |
ENST00000392580.5
|
B3GAT1
|
beta-1,3-glucuronyltransferase 1
|
|
chr22_+_43412008
Show fit
|
0.29 |
ENST00000334209.9
ENST00000443721.2
|
MPPED1
|
metallophosphoesterase domain containing 1
|
|
chr2_+_84971093
Show fit
|
0.29 |
ENST00000456682.1
ENST00000409785.9
|
KCMF1
|
potassium channel modulatory factor 1
|
|
chr8_+_135458027
Show fit
|
0.29 |
ENST00000524199.5
|
KHDRBS3
|
KH RNA binding domain containing, signal transduction associated 3
|
|
chr2_+_238426994
Show fit
|
0.29 |
ENST00000409297.1
|
ASB1
|
ankyrin repeat and SOCS box containing 1
|
|
chr9_+_135075422
Show fit
|
0.28 |
ENST00000371799.8
ENST00000277415.15
|
OLFM1
|
olfactomedin 1
|
|
chr12_+_120640552
Show fit
|
0.28 |
ENST00000316803.7
|
CABP1
|
calcium binding protein 1
|
|
chr6_-_90586883
Show fit
|
0.28 |
ENST00000369325.7
ENST00000369327.7
|
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7
|
|
chr4_+_70704713
Show fit
|
0.28 |
ENST00000417478.6
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr1_+_43389889
Show fit
|
0.28 |
ENST00000562955.2
ENST00000634258.3
|
SZT2
|
SZT2 subunit of KICSTOR complex
|
|
chr14_+_52552830
Show fit
|
0.28 |
ENST00000321662.11
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr1_-_1540350
Show fit
|
0.28 |
ENST00000624426.1
ENST00000425828.1
ENST00000378733.9
|
TMEM240
|
transmembrane protein 240
|
|
chr16_-_10942443
Show fit
|
0.28 |
ENST00000570440.2
ENST00000331808.5
|
DEXI
|
Dexi homolog
|
|
chr10_-_121598396
Show fit
|
0.28 |
ENST00000336553.10
ENST00000457416.6
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr3_-_53046031
Show fit
|
0.28 |
ENST00000482396.5
ENST00000394752.8
|
SFMBT1
|
Scm like with four mbt domains 1
|
|
chr1_-_160070102
Show fit
|
0.27 |
ENST00000638728.1
ENST00000637644.1
|
KCNJ10
|
potassium inwardly rectifying channel subfamily J member 10
|
|
chr2_+_235494024
Show fit
|
0.27 |
ENST00000304032.13
ENST00000409457.5
ENST00000336665.9
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr10_+_71212524
Show fit
|
0.27 |
ENST00000335350.10
|
UNC5B
|
unc-5 netrin receptor B
|
|
chr10_+_81875173
Show fit
|
0.27 |
ENST00000372141.7
ENST00000404547.5
|
NRG3
|
neuregulin 3
|
|
chr14_-_77320855
Show fit
|
0.27 |
ENST00000556394.2
ENST00000261534.9
|
POMT2
|
protein O-mannosyltransferase 2
|
|
chr15_+_92393841
Show fit
|
0.27 |
ENST00000268164.8
ENST00000539113.5
ENST00000555434.1
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2
|
|
chr14_+_73950252
Show fit
|
0.27 |
ENST00000629426.2
|
COQ6
|
coenzyme Q6, monooxygenase
|
|
chr1_-_150010675
Show fit
|
0.27 |
ENST00000417191.2
ENST00000581312.6
|
OTUD7B
|
OTU deubiquitinase 7B
|
|
chr9_+_35829217
Show fit
|
0.27 |
ENST00000439587.6
ENST00000643932.2
ENST00000650015.1
ENST00000377991.9
|
TMEM8B
|
transmembrane protein 8B
|
|
chr4_-_11428868
Show fit
|
0.27 |
ENST00000002596.6
|
HS3ST1
|
heparan sulfate-glucosamine 3-sulfotransferase 1
|
|
chr14_+_103385374
Show fit
|
0.27 |
ENST00000678179.1
ENST00000676938.1
ENST00000678619.1
ENST00000440884.7
ENST00000560417.6
ENST00000679330.1
ENST00000556744.2
ENST00000676897.1
ENST00000677560.1
ENST00000561314.6
ENST00000677829.1
ENST00000677133.1
ENST00000676645.1
ENST00000678175.1
ENST00000429436.7
ENST00000677360.1
ENST00000678237.1
ENST00000677347.1
ENST00000677432.1
|
MARK3
|
microtubule affinity regulating kinase 3
|
|
chr10_+_104641282
Show fit
|
0.27 |
ENST00000369701.8
|
SORCS3
|
sortilin related VPS10 domain containing receptor 3
|
|
chr10_+_103493931
Show fit
|
0.26 |
ENST00000369780.8
|
NEURL1
|
neuralized E3 ubiquitin protein ligase 1
|
|
chr3_-_48430045
Show fit
|
0.26 |
ENST00000296440.11
|
PLXNB1
|
plexin B1
|
|
chr5_+_133051983
Show fit
|
0.26 |
ENST00000304858.7
ENST00000617074.4
ENST00000615899.1
|
HSPA4
|
heat shock protein family A (Hsp70) member 4
|
|
chrX_-_63754664
Show fit
|
0.26 |
ENST00000677315.1
ENST00000636392.1
ENST00000637040.1
ENST00000637178.1
ENST00000637557.1
ENST00000636048.1
ENST00000638021.1
ENST00000672513.1
|
ENSG00000288661.1
ARHGEF9
|
novel protein
Cdc42 guanine nucleotide exchange factor 9
|
|
chr9_-_137221323
Show fit
|
0.26 |
ENST00000391553.2
ENST00000392827.2
|
RNF208
|
ring finger protein 208
|
|
chr6_-_90587018
Show fit
|
0.26 |
ENST00000369332.7
ENST00000369329.8
|
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7
|
|
chr3_+_196568611
Show fit
|
0.26 |
ENST00000440469.1
ENST00000311630.7
|
FBXO45
|
F-box protein 45
|
|
chr1_-_160262540
Show fit
|
0.25 |
ENST00000368074.6
|
DCAF8
|
DDB1 and CUL4 associated factor 8
|
|
chr2_+_109794263
Show fit
|
0.25 |
ENST00000272454.10
|
RGPD5
|
RANBP2 like and GRIP domain containing 5
|
|
chr7_+_45574358
Show fit
|
0.25 |
ENST00000297323.12
|
ADCY1
|
adenylate cyclase 1
|
|
chr4_-_139084191
Show fit
|
0.25 |
ENST00000512627.1
|
ELF2
|
E74 like ETS transcription factor 2
|
|
chr3_-_184017863
Show fit
|
0.25 |
ENST00000427120.6
ENST00000334444.11
ENST00000392579.6
ENST00000382494.6
ENST00000265586.10
ENST00000446941.2
|
ABCC5
|
ATP binding cassette subfamily C member 5
|
|
chr1_-_160070150
Show fit
|
0.25 |
ENST00000644903.1
|
KCNJ10
|
potassium inwardly rectifying channel subfamily J member 10
|
|
chr10_-_121598234
Show fit
|
0.25 |
ENST00000369058.7
ENST00000369060.8
ENST00000359354.6
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr11_-_110296605
Show fit
|
0.25 |
ENST00000534683.1
ENST00000647231.1
ENST00000645495.2
|
RDX
|
radixin
|
|
chr8_-_12755457
Show fit
|
0.25 |
ENST00000398246.8
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1
|
|
chr22_-_31346317
Show fit
|
0.25 |
ENST00000266269.10
|
PATZ1
|
POZ/BTB and AT hook containing zinc finger 1
|
|
chr1_-_160262407
Show fit
|
0.25 |
ENST00000419626.1
ENST00000610139.5
ENST00000475733.5
ENST00000407642.6
ENST00000368073.7
ENST00000326837.6
|
DCAF8
|
DDB1 and CUL4 associated factor 8
|
|
chr1_+_35557435
Show fit
|
0.25 |
ENST00000373253.7
|
NCDN
|
neurochondrin
|
|
chr16_+_74999003
Show fit
|
0.25 |
ENST00000335325.9
ENST00000320619.10
|
ZNRF1
|
zinc and ring finger 1
|
|
chr4_-_576278
Show fit
|
0.24 |
ENST00000610212.3
|
TMEM271
|
transmembrane protein 271
|
|
chr16_+_22814154
Show fit
|
0.24 |
ENST00000261374.4
|
HS3ST2
|
heparan sulfate-glucosamine 3-sulfotransferase 2
|
|
chr3_-_18445565
Show fit
|
0.24 |
ENST00000415069.5
|
SATB1
|
SATB homeobox 1
|
|
chr7_+_138460238
Show fit
|
0.24 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24
|
|
chr17_-_65561137
Show fit
|
0.24 |
ENST00000580513.1
|
AXIN2
|
axin 2
|
|
chr1_+_4654601
Show fit
|
0.24 |
ENST00000378191.5
|
AJAP1
|
adherens junctions associated protein 1
|
|
chr6_+_139028838
Show fit
|
0.24 |
ENST00000461027.1
|
ABRACL
|
ABRA C-terminal like
|
|
chr9_-_127854636
Show fit
|
0.24 |
ENST00000344849.4
ENST00000373203.9
|
ENG
|
endoglin
|
|
chr12_-_57430956
Show fit
|
0.24 |
ENST00000347140.7
ENST00000402412.5
|
R3HDM2
|
R3H domain containing 2
|
|
chr3_+_49554436
Show fit
|
0.24 |
ENST00000296452.5
|
BSN
|
bassoon presynaptic cytomatrix protein
|
|
chr13_+_31199959
Show fit
|
0.24 |
ENST00000343307.5
|
B3GLCT
|
beta 3-glucosyltransferase
|