Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
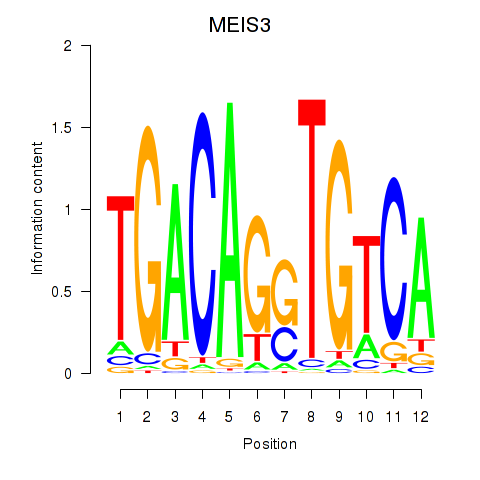
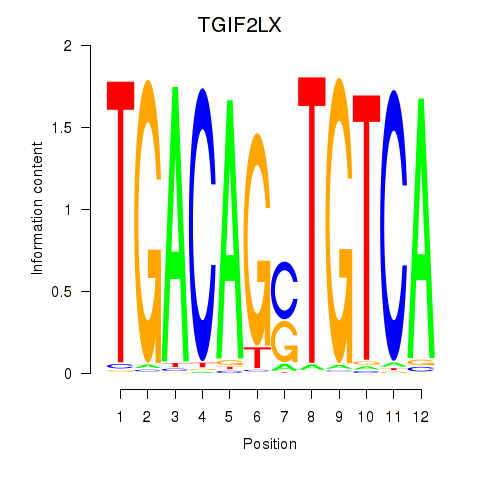
Results for MEIS3_TGIF2LX
Z-value: 1.03
Transcription factors associated with MEIS3_TGIF2LX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEIS3
|
ENSG00000105419.18 | Meis homeobox 3 |
|
TGIF2LX
|
ENSG00000153779.11 | TGFB induced factor homeobox 2 like X-linked |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TGIF2LX | hg38_v1_chrX_+_89921903_89921956 | 0.11 | 5.5e-01 | Click! |
| MEIS3 | hg38_v1_chr19_-_47419490_47419538 | -0.10 | 5.9e-01 | Click! |
Activity profile of MEIS3_TGIF2LX motif
Sorted Z-values of MEIS3_TGIF2LX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_47190013 | 4.29 |
ENST00000294338.7
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr17_+_42851167 | 3.80 |
ENST00000613571.1
ENST00000308423.7 |
AOC3
|
amine oxidase copper containing 3 |
| chr16_+_1240698 | 2.61 |
ENST00000561736.2
ENST00000338844.8 ENST00000461509.6 |
TPSAB1
|
tryptase alpha/beta 1 |
| chr16_-_1230089 | 2.56 |
ENST00000612142.1
ENST00000606293.5 |
TPSB2
|
tryptase beta 2 |
| chr11_-_107719657 | 2.56 |
ENST00000525934.1
ENST00000531293.1 |
SLN
|
sarcolipin |
| chr7_+_76510528 | 2.47 |
ENST00000334348.8
|
UPK3B
|
uroplakin 3B |
| chr7_-_27152561 | 2.37 |
ENST00000467897.2
ENST00000612286.5 ENST00000498652.1 |
ENSG00000273433.1
HOXA3
ENSG00000272801.1
|
novel transcript homeobox A3 novel transcript |
| chr16_+_29778252 | 2.25 |
ENST00000400752.6
|
ZG16
|
zymogen granule protein 16 |
| chr1_+_86468902 | 2.01 |
ENST00000394711.2
|
CLCA1
|
chloride channel accessory 1 |
| chr9_-_22009272 | 1.99 |
ENST00000380142.5
ENST00000276925.7 |
CDKN2B
|
cyclin dependent kinase inhibitor 2B |
| chr11_-_76669985 | 1.98 |
ENST00000407242.6
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr1_-_155192867 | 1.79 |
ENST00000342482.8
ENST00000343256.9 ENST00000368389.6 ENST00000368390.7 ENST00000368396.8 ENST00000368398.7 ENST00000471283.5 ENST00000337604.6 ENST00000368392.7 ENST00000368393.7 ENST00000438413.5 ENST00000457295.6 ENST00000462215.5 ENST00000620103.4 ENST00000338684.9 ENST00000610359.4 ENST00000611571.4 ENST00000611577.4 ENST00000612778.4 ENST00000614519.4 ENST00000615517.4 |
MUC1
|
mucin 1, cell surface associated |
| chr12_+_12891554 | 1.79 |
ENST00000014914.6
|
GPRC5A
|
G protein-coupled receptor class C group 5 member A |
| chr17_-_44268119 | 1.71 |
ENST00000399246.3
ENST00000262418.12 |
SLC4A1
|
solute carrier family 4 member 1 (Diego blood group) |
| chr11_-_119317119 | 1.64 |
ENST00000264036.6
|
MCAM
|
melanoma cell adhesion molecule |
| chr2_+_96536743 | 1.53 |
ENST00000673792.1
ENST00000357485.8 |
ARID5A
|
AT-rich interaction domain 5A |
| chr21_-_42315336 | 1.49 |
ENST00000398431.2
ENST00000518498.3 |
TFF3
|
trefoil factor 3 |
| chr2_+_96537254 | 1.49 |
ENST00000454558.2
|
ARID5A
|
AT-rich interaction domain 5A |
| chr17_-_35795592 | 1.43 |
ENST00000615136.4
ENST00000605424.6 ENST00000612672.1 |
MMP28
|
matrix metallopeptidase 28 |
| chr17_+_39626702 | 1.39 |
ENST00000580825.5
|
PPP1R1B
|
protein phosphatase 1 regulatory inhibitor subunit 1B |
| chr12_-_10849464 | 1.37 |
ENST00000544994.5
ENST00000228811.8 ENST00000540107.2 |
PRR4
|
proline rich 4 |
| chr16_+_66376341 | 1.36 |
ENST00000562048.5
ENST00000568155.2 |
CDH5
|
cadherin 5 |
| chr17_-_41624541 | 1.34 |
ENST00000540235.5
ENST00000311208.13 |
KRT17
|
keratin 17 |
| chr2_+_89913982 | 1.27 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr1_-_153544997 | 1.21 |
ENST00000368715.5
|
S100A4
|
S100 calcium binding protein A4 |
| chr9_-_33447553 | 1.21 |
ENST00000645858.1
ENST00000297991.6 |
AQP3
|
aquaporin 3 (Gill blood group) |
| chr16_+_1256059 | 1.18 |
ENST00000397534.6
ENST00000211076.5 |
TPSD1
|
tryptase delta 1 |
| chr19_-_51027662 | 1.18 |
ENST00000594768.5
|
KLK11
|
kallikrein related peptidase 11 |
| chr18_-_55322334 | 1.15 |
ENST00000630720.3
|
TCF4
|
transcription factor 4 |
| chr21_+_29130630 | 1.14 |
ENST00000399926.5
ENST00000399928.6 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr5_+_115962467 | 1.11 |
ENST00000357872.9
ENST00000395528.6 |
LVRN
|
laeverin |
| chr17_-_41624803 | 1.09 |
ENST00000463128.5
|
KRT17
|
keratin 17 |
| chr17_-_41624382 | 1.08 |
ENST00000577817.3
|
KRT17
|
keratin 17 |
| chr3_+_155083889 | 1.06 |
ENST00000680282.1
|
MME
|
membrane metalloendopeptidase |
| chr1_+_99646025 | 1.06 |
ENST00000263174.9
ENST00000605497.5 ENST00000615664.1 |
PALMD
|
palmdelphin |
| chr17_+_39626944 | 1.03 |
ENST00000582680.5
ENST00000254079.9 |
PPP1R1B
|
protein phosphatase 1 regulatory inhibitor subunit 1B |
| chr19_-_51026593 | 1.03 |
ENST00000600362.5
ENST00000453757.8 ENST00000601671.1 |
KLK11
|
kallikrein related peptidase 11 |
| chr11_+_18266254 | 1.01 |
ENST00000532858.5
ENST00000649195.1 ENST00000356524.9 ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr10_-_7619660 | 1.00 |
ENST00000613909.4
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain 5 |
| chr11_-_76670737 | 0.99 |
ENST00000260061.9
ENST00000404995.5 |
LRRC32
|
leucine rich repeat containing 32 |
| chr19_-_51028015 | 0.99 |
ENST00000319720.11
|
KLK11
|
kallikrein related peptidase 11 |
| chr13_+_77535742 | 0.92 |
ENST00000377246.7
|
SCEL
|
sciellin |
| chr13_+_77535681 | 0.92 |
ENST00000349847.4
|
SCEL
|
sciellin |
| chr13_+_77535669 | 0.90 |
ENST00000535157.5
|
SCEL
|
sciellin |
| chr3_+_155083523 | 0.90 |
ENST00000680057.1
|
MME
|
membrane metalloendopeptidase |
| chr5_-_170389349 | 0.89 |
ENST00000274629.9
|
KCNMB1
|
potassium calcium-activated channel subfamily M regulatory beta subunit 1 |
| chr9_-_112890868 | 0.88 |
ENST00000374228.5
|
SLC46A2
|
solute carrier family 46 member 2 |
| chr1_+_45913647 | 0.87 |
ENST00000674079.1
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr14_-_106627685 | 0.84 |
ENST00000390629.3
|
IGHV4-59
|
immunoglobulin heavy variable 4-59 |
| chr2_+_89862438 | 0.81 |
ENST00000448155.2
|
IGKV1D-39
|
immunoglobulin kappa variable 1D-39 |
| chr19_-_51027954 | 0.81 |
ENST00000391804.7
|
KLK11
|
kallikrein related peptidase 11 |
| chr15_+_65045377 | 0.79 |
ENST00000334287.3
|
SLC51B
|
solute carrier family 51 subunit beta |
| chr3_-_49029378 | 0.78 |
ENST00000442157.2
ENST00000326739.9 ENST00000677010.1 ENST00000678724.1 ENST00000429182.6 |
IMPDH2
|
inosine monophosphate dehydrogenase 2 |
| chrX_+_136536099 | 0.77 |
ENST00000440515.5
ENST00000456412.1 |
VGLL1
|
vestigial like family member 1 |
| chr2_+_90100235 | 0.76 |
ENST00000492446.1
|
IGKV1D-16
|
immunoglobulin kappa variable 1D-16 |
| chr19_+_1000419 | 0.73 |
ENST00000234389.3
|
GRIN3B
|
glutamate ionotropic receptor NMDA type subunit 3B |
| chrX_-_11427725 | 0.72 |
ENST00000380736.5
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr4_-_186596770 | 0.71 |
ENST00000512772.5
|
FAT1
|
FAT atypical cadherin 1 |
| chr3_-_134029914 | 0.71 |
ENST00000493729.5
ENST00000310926.11 |
SLCO2A1
|
solute carrier organic anion transporter family member 2A1 |
| chr5_+_151020438 | 0.71 |
ENST00000622181.4
ENST00000614343.4 ENST00000388825.9 ENST00000521650.5 ENST00000517973.1 |
GPX3
|
glutathione peroxidase 3 |
| chr11_-_35526024 | 0.70 |
ENST00000615849.4
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr4_-_18021727 | 0.70 |
ENST00000675605.1
ENST00000675927.1 ENST00000674942.1 ENST00000675143.1 ENST00000382226.5 ENST00000326877.8 ENST00000635767.1 |
LCORL
|
ligand dependent nuclear receptor corepressor like |
| chr11_+_1099730 | 0.68 |
ENST00000674892.1
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr9_-_92536645 | 0.68 |
ENST00000395534.2
|
ECM2
|
extracellular matrix protein 2 |
| chrX_-_13817027 | 0.65 |
ENST00000493677.5
ENST00000355135.6 ENST00000316715.9 |
GPM6B
|
glycoprotein M6B |
| chr1_-_183590596 | 0.63 |
ENST00000418089.5
ENST00000413720.5 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr21_+_29130961 | 0.63 |
ENST00000399925.5
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr19_+_18419374 | 0.63 |
ENST00000270061.12
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr11_+_102317492 | 0.62 |
ENST00000673846.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr9_+_34990250 | 0.62 |
ENST00000454002.6
ENST00000545841.5 |
DNAJB5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr13_-_38990824 | 0.61 |
ENST00000379631.9
|
STOML3
|
stomatin like 3 |
| chr9_-_83817632 | 0.60 |
ENST00000376365.7
ENST00000376371.7 |
GKAP1
|
G kinase anchoring protein 1 |
| chr19_+_676385 | 0.60 |
ENST00000166139.9
|
FSTL3
|
follistatin like 3 |
| chr7_+_76510516 | 0.60 |
ENST00000257632.9
|
UPK3B
|
uroplakin 3B |
| chr6_+_34236865 | 0.59 |
ENST00000674029.1
ENST00000447654.5 ENST00000347617.10 ENST00000401473.7 ENST00000311487.9 |
HMGA1
|
high mobility group AT-hook 1 |
| chr2_-_89117844 | 0.59 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr10_+_97589715 | 0.57 |
ENST00000370640.5
|
C10orf62
|
chromosome 10 open reading frame 62 |
| chr1_-_183590439 | 0.57 |
ENST00000367535.8
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr11_-_101129806 | 0.57 |
ENST00000325455.10
ENST00000617858.4 ENST00000619228.2 |
PGR
|
progesterone receptor |
| chr19_+_18419322 | 0.56 |
ENST00000348495.10
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr11_-_65557740 | 0.56 |
ENST00000526927.5
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr1_+_19596960 | 0.56 |
ENST00000617872.4
ENST00000322753.7 ENST00000602662.1 |
MICOS10
NBL1
|
mitochondrial contact site and cristae organizing system subunit 10 NBL1, DAN family BMP antagonist |
| chr11_+_102317542 | 0.55 |
ENST00000532808.5
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr12_+_104064520 | 0.54 |
ENST00000229330.9
|
HCFC2
|
host cell factor C2 |
| chr11_+_827545 | 0.53 |
ENST00000528542.6
|
CRACR2B
|
calcium release activated channel regulator 2B |
| chr1_-_201112420 | 0.52 |
ENST00000362061.4
ENST00000681874.1 |
CACNA1S
|
calcium voltage-gated channel subunit alpha1 S |
| chr19_+_47256518 | 0.51 |
ENST00000643617.1
ENST00000221922.11 |
CCDC9
|
coiled-coil domain containing 9 |
| chr1_-_31798755 | 0.51 |
ENST00000452755.6
|
SPOCD1
|
SPOC domain containing 1 |
| chr7_+_134646798 | 0.50 |
ENST00000418040.5
ENST00000393132.2 |
BPGM
|
bisphosphoglycerate mutase |
| chr1_+_45913583 | 0.50 |
ENST00000372008.6
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr9_+_132978687 | 0.50 |
ENST00000372122.4
ENST00000372123.5 |
GFI1B
|
growth factor independent 1B transcriptional repressor |
| chr18_+_3449817 | 0.50 |
ENST00000407501.6
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr13_-_77919459 | 0.49 |
ENST00000643890.1
|
EDNRB
|
endothelin receptor type B |
| chr22_+_22668286 | 0.49 |
ENST00000390304.2
|
IGLV3-27
|
immunoglobulin lambda variable 3-27 |
| chrX_+_23908006 | 0.49 |
ENST00000379211.8
ENST00000648352.1 |
CXorf58
|
chromosome X open reading frame 58 |
| chr13_-_38990856 | 0.49 |
ENST00000423210.1
|
STOML3
|
stomatin like 3 |
| chr1_+_81699665 | 0.48 |
ENST00000359929.7
|
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr1_-_183590876 | 0.47 |
ENST00000367536.5
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr2_-_217905445 | 0.47 |
ENST00000413554.5
|
TNS1
|
tensin 1 |
| chr3_-_64268161 | 0.47 |
ENST00000564377.6
|
PRICKLE2
|
prickle planar cell polarity protein 2 |
| chr7_+_76510608 | 0.47 |
ENST00000394849.1
|
UPK3B
|
uroplakin 3B |
| chr11_+_828150 | 0.46 |
ENST00000450448.5
|
CRACR2B
|
calcium release activated channel regulator 2B |
| chr11_+_68008542 | 0.46 |
ENST00000614849.4
|
ALDH3B1
|
aldehyde dehydrogenase 3 family member B1 |
| chr12_-_50025394 | 0.46 |
ENST00000454520.6
ENST00000546595.5 ENST00000548824.5 ENST00000549777.5 ENST00000546723.5 ENST00000427314.6 ENST00000552157.5 ENST00000552310.5 ENST00000548644.5 ENST00000546786.5 ENST00000550149.5 ENST00000546764.5 ENST00000552004.2 ENST00000548320.5 ENST00000312377.10 ENST00000547905.5 ENST00000550651.5 ENST00000551145.5 ENST00000552921.5 |
RACGAP1
|
Rac GTPase activating protein 1 |
| chr1_-_201112451 | 0.46 |
ENST00000367338.7
|
CACNA1S
|
calcium voltage-gated channel subunit alpha1 S |
| chr3_+_29280945 | 0.45 |
ENST00000434693.6
|
RBMS3
|
RNA binding motif single stranded interacting protein 3 |
| chr21_-_46323806 | 0.44 |
ENST00000397680.5
ENST00000445935.5 ENST00000291691.12 ENST00000397682.7 |
C21orf58
|
chromosome 21 open reading frame 58 |
| chr17_-_19745602 | 0.44 |
ENST00000444455.5
ENST00000439102.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr4_-_183320267 | 0.44 |
ENST00000323319.7
|
CLDN22
|
claudin 22 |
| chr18_+_3449620 | 0.44 |
ENST00000405385.7
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr19_+_18419425 | 0.44 |
ENST00000601357.6
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr3_-_12841527 | 0.43 |
ENST00000396953.6
ENST00000457131.1 ENST00000435983.5 ENST00000273223.10 ENST00000429711.7 ENST00000396957.5 |
RPL32
|
ribosomal protein L32 |
| chr7_+_134646845 | 0.43 |
ENST00000344924.8
|
BPGM
|
bisphosphoglycerate mutase |
| chr5_-_59586393 | 0.43 |
ENST00000505453.1
ENST00000360047.9 |
PDE4D
|
phosphodiesterase 4D |
| chrX_-_118116746 | 0.43 |
ENST00000371882.5
ENST00000545703.5 ENST00000540167.5 |
KLHL13
|
kelch like family member 13 |
| chr8_+_66043413 | 0.42 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr16_+_67326808 | 0.42 |
ENST00000329956.11
ENST00000569499.6 ENST00000561948.1 |
LRRC36
|
leucine rich repeat containing 36 |
| chr2_+_188991663 | 0.41 |
ENST00000450867.1
|
COL3A1
|
collagen type III alpha 1 chain |
| chr11_-_2149603 | 0.41 |
ENST00000643349.1
|
ENSG00000284779.2
|
novel protein |
| chr17_+_75093287 | 0.39 |
ENST00000538213.6
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 member 5 |
| chrX_+_17737443 | 0.39 |
ENST00000398080.5
ENST00000380045.7 ENST00000380043.7 ENST00000380041.8 |
SCML1
|
Scm polycomb group protein like 1 |
| chr3_-_187745460 | 0.39 |
ENST00000406870.7
|
BCL6
|
BCL6 transcription repressor |
| chr10_+_122211175 | 0.39 |
ENST00000496913.6
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr13_+_114179272 | 0.38 |
ENST00000646158.1
ENST00000636692.1 |
CFAP97D2
|
CFAP97 domain containing 2 |
| chr1_-_2003808 | 0.37 |
ENST00000493964.5
|
CFAP74
|
cilia and flagella associated protein 74 |
| chr12_-_25648544 | 0.37 |
ENST00000540106.5
ENST00000445693.5 ENST00000545543.1 |
LMNTD1
|
lamin tail domain containing 1 |
| chr2_+_24491860 | 0.37 |
ENST00000406961.5
ENST00000405141.5 |
NCOA1
|
nuclear receptor coactivator 1 |
| chr11_-_78417788 | 0.36 |
ENST00000361507.5
|
GAB2
|
GRB2 associated binding protein 2 |
| chr19_-_8981342 | 0.36 |
ENST00000397910.8
|
MUC16
|
mucin 16, cell surface associated |
| chr5_-_42887392 | 0.35 |
ENST00000514218.5
|
SELENOP
|
selenoprotein P |
| chr12_-_122730828 | 0.34 |
ENST00000432564.3
|
HCAR1
|
hydroxycarboxylic acid receptor 1 |
| chr3_+_131026844 | 0.34 |
ENST00000510769.5
ENST00000383366.9 ENST00000510688.5 ENST00000511262.5 |
NEK11
|
NIMA related kinase 11 |
| chr5_+_15500172 | 0.34 |
ENST00000504595.2
|
FBXL7
|
F-box and leucine rich repeat protein 7 |
| chr1_-_2003780 | 0.33 |
ENST00000682832.1
|
CFAP74
|
cilia and flagella associated protein 74 |
| chr16_+_20806424 | 0.33 |
ENST00000568894.5
|
REXO5
|
RNA exonuclease 5 |
| chr1_+_201739864 | 0.32 |
ENST00000367295.5
|
NAV1
|
neuron navigator 1 |
| chr13_+_114179247 | 0.32 |
ENST00000635901.2
|
CFAP97D2
|
CFAP97 domain containing 2 |
| chr10_-_87841336 | 0.32 |
ENST00000495903.1
|
ATAD1
|
ATPase family AAA domain containing 1 |
| chr3_-_71305986 | 0.32 |
ENST00000647614.1
|
FOXP1
|
forkhead box P1 |
| chr16_+_20806439 | 0.31 |
ENST00000568046.5
|
REXO5
|
RNA exonuclease 5 |
| chr5_+_102808057 | 0.31 |
ENST00000684043.1
ENST00000682407.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr13_-_98521998 | 0.31 |
ENST00000376547.7
|
STK24
|
serine/threonine kinase 24 |
| chr16_+_20806630 | 0.30 |
ENST00000568647.5
|
REXO5
|
RNA exonuclease 5 |
| chr21_-_34511317 | 0.30 |
ENST00000611936.1
ENST00000337385.7 |
KCNE1
|
potassium voltage-gated channel subfamily E regulatory subunit 1 |
| chr18_+_3449413 | 0.30 |
ENST00000549253.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr1_+_36156096 | 0.30 |
ENST00000474796.2
ENST00000373150.8 ENST00000373151.6 |
MAP7D1
|
MAP7 domain containing 1 |
| chr11_+_47586678 | 0.29 |
ENST00000538490.3
|
FAM180B
|
family with sequence similarity 180 member B |
| chr9_+_132978651 | 0.29 |
ENST00000636137.1
|
GFI1B
|
growth factor independent 1B transcriptional repressor |
| chr2_+_227325349 | 0.29 |
ENST00000436237.5
ENST00000443428.6 ENST00000418961.5 |
MFF
|
mitochondrial fission factor |
| chr8_+_26383043 | 0.28 |
ENST00000380629.7
|
BNIP3L
|
BCL2 interacting protein 3 like |
| chr6_-_30160880 | 0.28 |
ENST00000376704.3
|
TRIM10
|
tripartite motif containing 10 |
| chr17_+_60677822 | 0.27 |
ENST00000407086.8
ENST00000589222.5 ENST00000626960.2 ENST00000390652.9 |
BCAS3
|
BCAS3 microtubule associated cell migration factor |
| chr1_-_22143088 | 0.27 |
ENST00000290167.11
|
WNT4
|
Wnt family member 4 |
| chr11_-_842508 | 0.27 |
ENST00000322028.5
|
POLR2L
|
RNA polymerase II, I and III subunit L |
| chr16_+_20806517 | 0.26 |
ENST00000348433.10
ENST00000568501.5 ENST00000261377.11 ENST00000566276.5 |
REXO5
|
RNA exonuclease 5 |
| chr1_-_51325924 | 0.26 |
ENST00000532836.5
ENST00000422925.6 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr1_+_168280872 | 0.26 |
ENST00000367821.8
|
TBX19
|
T-box transcription factor 19 |
| chr17_+_36103819 | 0.25 |
ENST00000615863.2
ENST00000621626.1 |
CCL4
|
C-C motif chemokine ligand 4 |
| chr16_+_20806604 | 0.25 |
ENST00000565340.5
|
REXO5
|
RNA exonuclease 5 |
| chr22_+_38656627 | 0.25 |
ENST00000411557.5
ENST00000396811.6 ENST00000216029.7 ENST00000416285.5 |
CBY1
|
chibby family member 1, beta catenin antagonist |
| chr17_+_7306975 | 0.24 |
ENST00000336452.11
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr7_-_142258027 | 0.24 |
ENST00000552471.1
ENST00000547058.6 |
PRSS58
|
serine protease 58 |
| chr1_-_31798648 | 0.24 |
ENST00000528579.1
|
SPOCD1
|
SPOC domain containing 1 |
| chr19_-_39335999 | 0.23 |
ENST00000602185.5
ENST00000598034.5 ENST00000601387.5 ENST00000595636.1 ENST00000253054.12 ENST00000594700.5 ENST00000597595.6 |
GMFG
|
glia maturation factor gamma |
| chr19_-_49896868 | 0.23 |
ENST00000593956.5
ENST00000391826.7 |
IL4I1
|
interleukin 4 induced 1 |
| chr2_+_161231078 | 0.23 |
ENST00000439442.1
|
TANK
|
TRAF family member associated NFKB activator |
| chr19_-_45322867 | 0.23 |
ENST00000221476.4
|
CKM
|
creatine kinase, M-type |
| chr10_+_28533706 | 0.23 |
ENST00000442148.6
ENST00000448193.5 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr3_-_122793772 | 0.22 |
ENST00000306103.3
|
HSPBAP1
|
HSPB1 associated protein 1 |
| chr2_+_231592858 | 0.22 |
ENST00000313965.4
|
TEX44
|
testis expressed 44 |
| chr3_+_178807293 | 0.22 |
ENST00000358316.7
ENST00000617329.1 |
KCNMB2
|
potassium calcium-activated channel subfamily M regulatory beta subunit 2 |
| chr22_+_21015027 | 0.22 |
ENST00000413302.6
ENST00000401443.5 |
P2RX6
|
purinergic receptor P2X 6 |
| chr3_-_53347512 | 0.21 |
ENST00000559748.5
ENST00000294241.10 ENST00000610213.6 ENST00000560624.5 |
DCP1A
|
decapping mRNA 1A |
| chr6_-_30161200 | 0.21 |
ENST00000449742.7
|
TRIM10
|
tripartite motif containing 10 |
| chr16_+_20806698 | 0.21 |
ENST00000564274.5
ENST00000563068.1 |
REXO5
|
RNA exonuclease 5 |
| chr17_+_8413124 | 0.21 |
ENST00000582812.5
|
NDEL1
|
nudE neurodevelopment protein 1 like 1 |
| chr3_-_71306012 | 0.20 |
ENST00000649431.1
ENST00000610810.5 |
FOXP1
|
forkhead box P1 |
| chr9_+_93263948 | 0.18 |
ENST00000448251.5
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr2_+_232406836 | 0.18 |
ENST00000295453.8
|
ALPG
|
alkaline phosphatase, germ cell |
| chrX_+_102307022 | 0.18 |
ENST00000604790.2
|
NXF2
|
nuclear RNA export factor 2 |
| chr5_-_16738341 | 0.18 |
ENST00000515803.5
|
MYO10
|
myosin X |
| chr19_-_10116827 | 0.17 |
ENST00000593054.5
|
EIF3G
|
eukaryotic translation initiation factor 3 subunit G |
| chr9_-_127778659 | 0.17 |
ENST00000314830.13
|
SH2D3C
|
SH2 domain containing 3C |
| chr1_-_145885826 | 0.17 |
ENST00000544626.2
ENST00000355594.9 |
ANKRD35
|
ankyrin repeat domain 35 |
| chrX_-_154547546 | 0.17 |
ENST00000440967.5
ENST00000369620.6 ENST00000393564.6 |
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr3_+_182793478 | 0.17 |
ENST00000493826.1
ENST00000323116.10 |
ATP11B
|
ATPase phospholipid transporting 11B (putative) |
| chr19_+_5455409 | 0.16 |
ENST00000222033.6
|
ZNRF4
|
zinc and ring finger 4 |
| chr1_-_54406385 | 0.16 |
ENST00000610401.5
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr17_-_5111836 | 0.16 |
ENST00000575898.5
|
ZNF232
|
zinc finger protein 232 |
| chr20_-_58515382 | 0.16 |
ENST00000371149.8
|
APCDD1L
|
APC down-regulated 1 like |
| chr7_+_142740206 | 0.16 |
ENST00000422143.2
|
TRBV29-1
|
T cell receptor beta variable 29-1 |
| chr2_+_23927309 | 0.15 |
ENST00000404924.5
|
UBXN2A
|
UBX domain protein 2A |
| chrX_+_40623423 | 0.15 |
ENST00000423387.4
|
MPC1L
|
mitochondrial pyruvate carrier 1 like |
| chr8_-_16567362 | 0.15 |
ENST00000518026.5
|
MSR1
|
macrophage scavenger receptor 1 |
| chr17_+_41689862 | 0.15 |
ENST00000586699.1
|
EIF1
|
eukaryotic translation initiation factor 1 |
| chr10_-_103452356 | 0.14 |
ENST00000260743.10
|
CALHM2
|
calcium homeostasis modulator family member 2 |
| chr10_+_102743938 | 0.14 |
ENST00000448841.7
|
WBP1L
|
WW domain binding protein 1 like |
| chr15_+_63121818 | 0.14 |
ENST00000413507.3
|
LACTB
|
lactamase beta |
| chr8_+_26383102 | 0.13 |
ENST00000523949.5
|
BNIP3L
|
BCL2 interacting protein 3 like |
| chr7_+_69967464 | 0.13 |
ENST00000664521.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEIS3_TGIF2LX
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.6 | 3.8 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.3 | 2.4 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 1.4 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.2 | 1.2 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.2 | 0.9 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.2 | 2.4 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.2 | 0.8 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.2 | 3.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.2 | 0.9 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.2 | 2.0 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.8 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 1.8 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 0.4 | GO:2001038 | regulation of cellular response to drug(GO:2001038) |
| 0.1 | 3.5 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.1 | 0.6 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 1.6 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 0.5 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.6 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.6 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 1.4 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.7 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 0.5 | GO:0035645 | enteric smooth muscle cell differentiation(GO:0035645) |
| 0.1 | 0.4 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 1.4 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 1.2 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.1 | 1.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.0 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 0.2 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.2 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.8 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 2.0 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.3 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.2 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 1.0 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.3 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 3.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 1.7 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.4 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 4.7 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 1.0 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.5 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 1.0 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.6 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.5 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 1.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0035853 | chromosome passenger complex localization to spindle midzone(GO:0035853) |
| 0.0 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0090071 | rhythmic synaptic transmission(GO:0060024) negative regulation of ribosome biogenesis(GO:0090071) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.4 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.1 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.2 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 1.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:0070091 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.0 | 1.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.7 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.5 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.4 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.1 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.0 | 0.4 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 2.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 0.7 | GO:0070701 | mucus layer(GO:0070701) |
| 0.2 | 1.7 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.5 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 1.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 2.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 2.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 3.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 3.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 2.0 | GO:0005903 | brush border(GO:0005903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.6 | 2.4 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.4 | 3.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 0.9 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.2 | 1.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.2 | 0.8 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.7 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.4 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 1.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.5 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 2.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.3 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.2 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 0.5 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 1.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 2.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 2.9 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.9 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 1.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.2 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 2.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 1.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.7 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 1.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 13.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.7 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 1.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.6 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 3.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 3.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.4 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 3.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 3.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 2.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.6 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 3.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 2.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |