Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for MYB
Z-value: 3.74
Transcription factors associated with MYB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYB
|
ENSG00000118513.19 | MYB proto-oncogene, transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYB | hg38_v1_chr6_+_135181361_135181514 | 0.37 | 3.7e-02 | Click! |
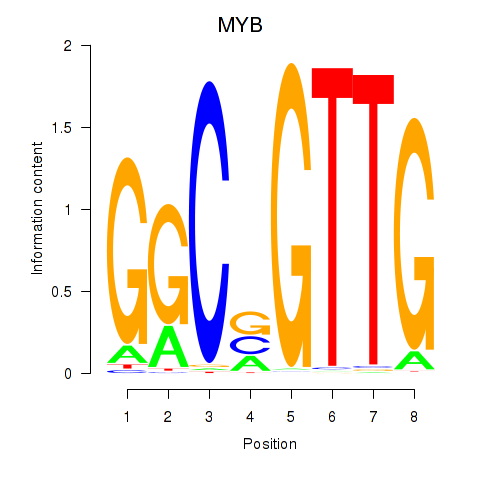
Activity profile of MYB motif
Sorted Z-values of MYB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYB
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 23.1 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 3.3 | 13.3 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 3.3 | 16.6 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 3.1 | 9.4 | GO:0009386 | translational attenuation(GO:0009386) |
| 3.0 | 9.0 | GO:0007309 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 2.7 | 56.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 2.0 | 6.0 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 1.5 | 6.0 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 1.4 | 9.7 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 1.3 | 5.3 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 1.3 | 3.9 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 1.3 | 10.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.2 | 8.3 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 1.2 | 7.1 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 1.1 | 3.3 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 1.1 | 5.5 | GO:0007538 | primary sex determination(GO:0007538) |
| 1.1 | 5.5 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 1.1 | 6.4 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 1.1 | 2.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 1.0 | 11.1 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 1.0 | 7.0 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 1.0 | 3.0 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 1.0 | 5.8 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.9 | 5.7 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.9 | 5.2 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.9 | 3.4 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.8 | 7.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.8 | 5.5 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.8 | 17.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.8 | 7.6 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.7 | 8.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.7 | 10.0 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.7 | 2.8 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.7 | 4.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.7 | 2.7 | GO:0009631 | cold acclimation(GO:0009631) |
| 0.7 | 2.7 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.6 | 3.9 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.6 | 17.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.6 | 1.3 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.6 | 9.4 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.6 | 3.6 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.6 | 3.6 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.6 | 1.8 | GO:0034963 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.6 | 1.8 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.6 | 1.7 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.6 | 1.7 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.5 | 2.2 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.5 | 2.7 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.5 | 1.6 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.5 | 2.7 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.5 | 2.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.5 | 2.5 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.5 | 3.5 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.5 | 7.0 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.5 | 3.5 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.5 | 1.0 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.5 | 1.4 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.5 | 3.8 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.5 | 1.9 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.4 | 2.6 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.4 | 2.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.4 | 11.0 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.4 | 4.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.4 | 11.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.4 | 1.6 | GO:1900229 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.4 | 72.9 | GO:0007286 | spermatid development(GO:0007286) |
| 0.4 | 1.2 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.4 | 4.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 3.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.4 | 2.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 3.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.4 | 10.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.4 | 1.4 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.4 | 7.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.4 | 5.0 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.4 | 1.4 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.3 | 2.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.3 | 2.4 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.3 | 3.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.3 | 1.7 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.3 | 1.3 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.3 | 1.0 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.3 | 1.3 | GO:0061015 | snRNA import into nucleus(GO:0061015) |
| 0.3 | 7.4 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.3 | 1.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.3 | 2.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 0.9 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.3 | 11.2 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.3 | 0.9 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.3 | 1.7 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.3 | 3.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.3 | 1.4 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.3 | 8.8 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.3 | 1.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.3 | 0.8 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.3 | 12.7 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.3 | 0.8 | GO:1901859 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.2 | 1.0 | GO:0035470 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) positive regulation of vascular wound healing(GO:0035470) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.2 | 5.7 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 1.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.2 | 0.7 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.2 | 5.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 0.7 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.2 | 3.9 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.2 | 11.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 0.7 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.2 | 0.9 | GO:2000850 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.2 | 13.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 15.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.2 | 0.9 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.2 | 1.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.2 | 1.9 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.2 | 2.1 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.2 | 0.8 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 3.9 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.2 | 2.2 | GO:0003341 | cilium movement(GO:0003341) |
| 0.2 | 3.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 8.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 2.0 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.2 | 4.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.2 | 1.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 2.9 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.2 | 1.3 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.2 | 2.1 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.2 | 0.9 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.2 | 1.7 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.2 | 0.5 | GO:1904247 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.2 | 1.4 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.2 | 4.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.2 | 94.6 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.2 | 0.9 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.2 | 5.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.2 | 1.6 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 2.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 2.4 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.2 | 0.7 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.2 | 4.2 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.2 | 1.2 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.2 | 1.0 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.2 | 9.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.2 | 1.5 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.6 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 4.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.2 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.1 | 2.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 8.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 2.2 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 0.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.9 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 2.9 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 0.5 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 14.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.1 | 5.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.6 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 1.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 2.9 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.4 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.1 | 1.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.8 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.7 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 3.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 3.7 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 2.7 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 2.0 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 0.5 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 2.6 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 3.1 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 2.6 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 1.6 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.9 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.1 | 2.0 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 4.0 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 3.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.8 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.7 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.6 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.6 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.4 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.1 | 1.9 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 1.7 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.1 | 5.7 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 2.5 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.4 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.5 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.5 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 4.0 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 2.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.4 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.5 | GO:1901673 | regulation of spindle assembly(GO:0090169) regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 7.1 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.3 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.1 | 0.3 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 0.8 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 1.6 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 7.8 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 1.7 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 10.6 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 4.0 | GO:1902850 | microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.1 | 0.4 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 1.1 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.3 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 0.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.2 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.9 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 1.2 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.2 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 0.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.6 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0002357 | defense response to tumor cell(GO:0002357) |
| 0.0 | 1.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 1.2 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 1.9 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.6 | GO:0060026 | convergent extension(GO:0060026) |
| 0.0 | 0.8 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 1.4 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.8 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 1.7 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.8 | GO:1900116 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.4 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 1.9 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.8 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 1.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.7 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 1.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 2.0 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 1.1 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 2.2 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 2.4 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.6 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 3.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.5 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.9 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 1.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.3 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.5 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 1.6 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.0 | 0.2 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.9 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 0.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 1.5 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.2 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 1.0 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.5 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.7 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.6 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.8 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 1.1 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 1.6 | GO:0032760 | positive regulation of tumor necrosis factor production(GO:0032760) |
| 0.0 | 3.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.3 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.6 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 2.2 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.0 | GO:0048561 | establishment of organ orientation(GO:0048561) |
| 0.0 | 0.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.2 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.3 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.6 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 1.1 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.1 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.2 | GO:0030849 | autosome(GO:0030849) |
| 1.9 | 3.8 | GO:1990923 | PET complex(GO:1990923) |
| 1.9 | 5.7 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 1.6 | 14.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.6 | 14.1 | GO:0071546 | pi-body(GO:0071546) |
| 1.5 | 3.1 | GO:0071547 | piP-body(GO:0071547) |
| 1.4 | 5.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.4 | 6.8 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.1 | 19.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 1.1 | 9.8 | GO:0098536 | deuterosome(GO:0098536) |
| 1.0 | 8.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.9 | 9.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.9 | 4.5 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.9 | 3.4 | GO:0030892 | mitotic cohesin complex(GO:0030892) |
| 0.8 | 10.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.8 | 9.6 | GO:0045495 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.7 | 2.2 | GO:0001534 | radial spoke(GO:0001534) |
| 0.7 | 3.0 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.7 | 8.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.6 | 3.5 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.6 | 14.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.6 | 1.7 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.5 | 34.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.5 | 1.4 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.5 | 5.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.5 | 4.7 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.5 | 2.8 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.4 | 9.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.4 | 1.5 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.4 | 18.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 13.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 2.7 | GO:0002177 | manchette(GO:0002177) |
| 0.3 | 8.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.3 | 1.0 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.3 | 4.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.3 | 5.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.3 | 5.3 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.3 | 46.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.3 | 1.6 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.3 | 0.8 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.3 | 1.6 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.3 | 33.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.3 | 9.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.2 | 0.7 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.2 | 8.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 1.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 2.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 1.9 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.2 | 2.5 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.2 | 8.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 4.6 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 0.8 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.2 | 1.0 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.2 | 0.8 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.2 | 3.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 4.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.2 | 10.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 2.6 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 1.0 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.2 | 16.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 3.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 7.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 3.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.7 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 4.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 3.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 4.1 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 1.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.8 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.7 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 2.0 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.6 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 7.5 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 3.9 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 0.6 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.9 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.8 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 1.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.7 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 0.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 2.0 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 5.7 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 3.1 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 0.3 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 11.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 13.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.0 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 2.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 12.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 1.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 2.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 3.7 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 9.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.3 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 4.3 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 5.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 2.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 2.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 3.0 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.5 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.2 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 1.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.4 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 8.8 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 1.6 | 6.3 | GO:0004040 | amidase activity(GO:0004040) |
| 1.4 | 15.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 1.2 | 7.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 1.1 | 3.4 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 1.1 | 5.5 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.0 | 9.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 1.0 | 9.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 1.0 | 3.0 | GO:0090422 | thiamine pyrophosphate transporter activity(GO:0090422) |
| 1.0 | 4.8 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 1.0 | 24.8 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.9 | 4.5 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.9 | 6.0 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.8 | 2.5 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.8 | 3.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.8 | 5.3 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.7 | 7.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.6 | 1.8 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.6 | 18.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.5 | 2.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.5 | 4.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.5 | 1.4 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.5 | 23.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.5 | 13.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.4 | 20.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.4 | 1.7 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.4 | 2.9 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.4 | 2.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.4 | 6.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.4 | 1.2 | GO:0052858 | peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) |
| 0.4 | 3.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.4 | 1.4 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.3 | 4.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 1.4 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.3 | 0.8 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.3 | 1.4 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.3 | 6.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.3 | 3.9 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.3 | 11.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.3 | 3.8 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.3 | 1.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.3 | 16.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.3 | 4.8 | GO:0019864 | IgG binding(GO:0019864) |
| 0.3 | 0.8 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.3 | 2.8 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.2 | 4.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.2 | 1.4 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.2 | 0.7 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.2 | 8.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.2 | 11.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 7.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.2 | 0.9 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.2 | 1.0 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.2 | 1.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 1.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 2.4 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 3.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 0.6 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.2 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 16.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 2.9 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.2 | 1.6 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 2.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 2.4 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.2 | 2.7 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 1.6 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 2.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 3.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 1.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 1.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 5.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 3.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 3.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.8 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 3.6 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 12.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 6.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 20.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.3 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.1 | 0.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.5 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 0.4 | GO:0030379 | neurotensin receptor activity, non-G-protein coupled(GO:0030379) |
| 0.1 | 1.6 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 3.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 4.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 6.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 2.9 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.5 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.3 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 2.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 14.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 3.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.6 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 2.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.8 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 7.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 4.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.9 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.6 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 7.2 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.3 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.1 | 2.0 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 9.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 1.6 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.7 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 4.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 3.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.8 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 3.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 2.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.8 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 1.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 19.4 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 8.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 0.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 2.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 7.1 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 6.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 5.5 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 1.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 10.7 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.3 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 5.7 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 1.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 1.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 2.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 5.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 1.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.8 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 3.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 49.4 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.5 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 3.2 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 2.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 12.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 1.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 2.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.7 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.0 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 2.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 18.6 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 0.0 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 47.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 16.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 17.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 1.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 1.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 9.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 11.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 7.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 6.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 4.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 9.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 13.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 8.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 2.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 2.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.0 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.4 | 6.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 7.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.4 | 16.6 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.3 | 27.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.3 | 3.8 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.2 | 5.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 6.0 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.2 | 25.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 4.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.2 | 2.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.1 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 0.3 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 6.5 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 6.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 3.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.9 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 4.3 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 6.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 4.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 3.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 8.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 10.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 4.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 5.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 1.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 2.2 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 2.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 2.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 5.6 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 3.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |