Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
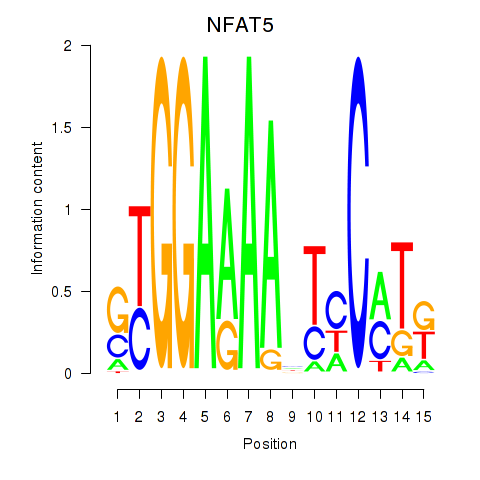
Results for NFAT5
Z-value: 0.70
Transcription factors associated with NFAT5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFAT5
|
ENSG00000102908.22 | nuclear factor of activated T cells 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFAT5 | hg38_v1_chr16_+_69566314_69566345 | 0.44 | 1.1e-02 | Click! |
Activity profile of NFAT5 motif
Sorted Z-values of NFAT5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_111923722 | 2.54 |
ENST00000527950.5
|
CRYAB
|
crystallin alpha B |
| chr12_+_101594849 | 2.34 |
ENST00000547405.5
ENST00000452455.6 ENST00000392934.7 ENST00000547509.5 ENST00000361685.6 ENST00000549145.5 ENST00000361466.7 ENST00000553190.5 ENST00000545503.6 ENST00000536007.5 ENST00000541119.5 ENST00000551300.5 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C1 |
| chr10_+_3104676 | 2.08 |
ENST00000415005.6
ENST00000468050.1 |
PFKP
|
phosphofructokinase, platelet |
| chr11_+_131370478 | 1.84 |
ENST00000374791.7
ENST00000683400.1 ENST00000436745.5 |
NTM
|
neurotrimin |
| chr1_+_159204860 | 1.82 |
ENST00000368122.4
ENST00000368121.6 |
ACKR1
|
atypical chemokine receptor 1 (Duffy blood group) |
| chr13_-_67230377 | 1.61 |
ENST00000544246.5
ENST00000377861.4 |
PCDH9
|
protocadherin 9 |
| chr22_-_37984534 | 1.59 |
ENST00000396884.8
|
SOX10
|
SRY-box transcription factor 10 |
| chr13_-_67230313 | 1.50 |
ENST00000377865.7
|
PCDH9
|
protocadherin 9 |
| chr6_-_53665748 | 1.45 |
ENST00000370905.4
|
KLHL31
|
kelch like family member 31 |
| chr6_+_152750789 | 1.44 |
ENST00000367244.8
ENST00000367243.7 |
VIP
|
vasoactive intestinal peptide |
| chr2_-_160200251 | 1.27 |
ENST00000428609.6
ENST00000409967.6 ENST00000283249.7 |
ITGB6
|
integrin subunit beta 6 |
| chr15_+_43593054 | 1.24 |
ENST00000453782.5
ENST00000300283.10 ENST00000437924.5 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr1_-_201377652 | 1.23 |
ENST00000455702.6
ENST00000367320.6 ENST00000421663.6 ENST00000412633.3 ENST00000367318.10 ENST00000422165.6 ENST00000656932.1 ENST00000438742.6 ENST00000367322.6 ENST00000458432.6 ENST00000509001.5 ENST00000660295.1 |
TNNT2
|
troponin T2, cardiac type |
| chr19_-_49155130 | 1.13 |
ENST00000595625.1
|
HRC
|
histidine rich calcium binding protein |
| chr3_-_116445458 | 1.10 |
ENST00000490035.7
|
LSAMP
|
limbic system associated membrane protein |
| chr6_+_150143018 | 1.06 |
ENST00000361131.5
|
PPP1R14C
|
protein phosphatase 1 regulatory inhibitor subunit 14C |
| chr2_-_160200310 | 1.03 |
ENST00000620391.4
|
ITGB6
|
integrin subunit beta 6 |
| chr4_+_119027335 | 1.02 |
ENST00000627783.2
|
SYNPO2
|
synaptopodin 2 |
| chr14_+_91114026 | 1.01 |
ENST00000521081.5
ENST00000520328.5 ENST00000524232.5 ENST00000522170.5 ENST00000256324.15 ENST00000519950.5 ENST00000523879.5 ENST00000521077.6 ENST00000518665.6 |
DGLUCY
|
D-glutamate cyclase |
| chr11_+_131911396 | 1.01 |
ENST00000425719.6
ENST00000374784.5 |
NTM
|
neurotrimin |
| chr3_-_116444983 | 0.94 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr10_+_6163818 | 0.93 |
ENST00000639949.1
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr19_-_45782479 | 0.93 |
ENST00000447742.6
ENST00000354227.9 ENST00000291270.9 ENST00000683086.1 ENST00000343373.10 |
DMPK
|
DM1 protein kinase |
| chr5_+_149581368 | 0.90 |
ENST00000333677.7
|
ARHGEF37
|
Rho guanine nucleotide exchange factor 37 |
| chr11_+_73648674 | 0.87 |
ENST00000535129.5
|
PLEKHB1
|
pleckstrin homology domain containing B1 |
| chrX_-_45200828 | 0.85 |
ENST00000398000.7
|
DIPK2B
|
divergent protein kinase domain 2B |
| chr9_+_68356603 | 0.84 |
ENST00000396396.6
|
PGM5
|
phosphoglucomutase 5 |
| chr14_-_25010604 | 0.84 |
ENST00000550887.5
|
STXBP6
|
syntaxin binding protein 6 |
| chr18_-_36122110 | 0.83 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 member 6 |
| chr20_+_36091409 | 0.73 |
ENST00000202028.9
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr17_+_34356472 | 0.73 |
ENST00000225844.7
|
CCL13
|
C-C motif chemokine ligand 13 |
| chr20_+_34516394 | 0.73 |
ENST00000357156.7
ENST00000300469.13 ENST00000374846.3 |
DYNLRB1
|
dynein light chain roadblock-type 1 |
| chr19_+_41708585 | 0.71 |
ENST00000398599.8
ENST00000221992.11 |
CEACAM5
|
CEA cell adhesion molecule 5 |
| chr19_+_859654 | 0.71 |
ENST00000592860.2
ENST00000327726.11 |
CFD
|
complement factor D |
| chr5_-_79512794 | 0.69 |
ENST00000282260.10
ENST00000508576.5 ENST00000535690.1 |
HOMER1
|
homer scaffold protein 1 |
| chr17_+_9842469 | 0.68 |
ENST00000304773.5
|
GLP2R
|
glucagon like peptide 2 receptor |
| chr7_+_149872955 | 0.66 |
ENST00000421974.7
ENST00000456496.7 |
ATP6V0E2
|
ATPase H+ transporting V0 subunit e2 |
| chr18_+_10454584 | 0.66 |
ENST00000355285.10
|
APCDD1
|
APC down-regulated 1 |
| chr12_+_26121984 | 0.65 |
ENST00000538142.5
|
SSPN
|
sarcospan |
| chr3_+_69762703 | 0.64 |
ENST00000433517.5
|
MITF
|
melanocyte inducing transcription factor |
| chr10_-_67696115 | 0.63 |
ENST00000433211.7
|
CTNNA3
|
catenin alpha 3 |
| chr14_+_91114431 | 0.62 |
ENST00000428926.6
ENST00000517362.5 |
DGLUCY
|
D-glutamate cyclase |
| chr6_+_116511626 | 0.62 |
ENST00000368599.4
|
CALHM5
|
calcium homeostasis modulator family member 5 |
| chr1_-_114757958 | 0.61 |
ENST00000525132.1
|
CSDE1
|
cold shock domain containing E1 |
| chr14_-_59630806 | 0.61 |
ENST00000342503.8
|
RTN1
|
reticulon 1 |
| chr15_+_48878070 | 0.60 |
ENST00000530028.3
ENST00000560490.1 |
EID1
|
EP300 interacting inhibitor of differentiation 1 |
| chr14_+_91114388 | 0.60 |
ENST00000519019.5
ENST00000523816.5 ENST00000517518.5 |
DGLUCY
|
D-glutamate cyclase |
| chrX_-_57137523 | 0.60 |
ENST00000614076.1
ENST00000374906.3 |
SPIN2A
|
spindlin family member 2A |
| chr14_+_91114667 | 0.60 |
ENST00000523894.5
ENST00000522322.5 ENST00000523771.5 |
DGLUCY
|
D-glutamate cyclase |
| chr9_+_136807911 | 0.59 |
ENST00000371671.9
ENST00000311502.12 ENST00000371663.10 |
RABL6
|
RAB, member RAS oncogene family like 6 |
| chr1_+_111346590 | 0.59 |
ENST00000369737.4
ENST00000369738.9 |
PIFO
|
primary cilia formation |
| chr1_+_11654368 | 0.57 |
ENST00000251546.8
|
FBXO44
|
F-box protein 44 |
| chr14_+_91114364 | 0.56 |
ENST00000518868.5
|
DGLUCY
|
D-glutamate cyclase |
| chr9_-_27005659 | 0.56 |
ENST00000380055.6
|
LRRC19
|
leucine rich repeat containing 19 |
| chr17_+_70075215 | 0.56 |
ENST00000283936.5
ENST00000615244.4 ENST00000392671.6 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr1_-_114757971 | 0.56 |
ENST00000261443.9
ENST00000534699.5 ENST00000339438.10 ENST00000358528.9 ENST00000529046.5 ENST00000525970.5 ENST00000369530.5 ENST00000530886.5 ENST00000610726.4 |
CSDE1
|
cold shock domain containing E1 |
| chr11_+_12377524 | 0.55 |
ENST00000334956.15
|
PARVA
|
parvin alpha |
| chr16_+_89921851 | 0.54 |
ENST00000554444.5
ENST00000556565.5 |
TUBB3
|
tubulin beta 3 class III |
| chr17_+_4950147 | 0.54 |
ENST00000522301.5
|
ENO3
|
enolase 3 |
| chr3_-_18445565 | 0.54 |
ENST00000415069.5
|
SATB1
|
SATB homeobox 1 |
| chr3_+_8501807 | 0.54 |
ENST00000426878.2
ENST00000397386.7 ENST00000415597.5 ENST00000157600.8 |
LMCD1
|
LIM and cysteine rich domains 1 |
| chr2_+_130963642 | 0.53 |
ENST00000409303.6
|
ARHGEF4
|
Rho guanine nucleotide exchange factor 4 |
| chrX_+_136497586 | 0.52 |
ENST00000218364.5
|
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr19_-_45782388 | 0.51 |
ENST00000458663.6
|
DMPK
|
DM1 protein kinase |
| chr22_-_29315656 | 0.51 |
ENST00000401450.3
ENST00000216101.7 |
RASL10A
|
RAS like family 10 member A |
| chr5_+_140806929 | 0.50 |
ENST00000378125.4
ENST00000618834.1 ENST00000530339.2 ENST00000512229.6 ENST00000672575.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr2_+_48568981 | 0.50 |
ENST00000394754.5
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr19_-_54364908 | 0.48 |
ENST00000391742.7
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1 |
| chr2_-_160200289 | 0.46 |
ENST00000409872.1
|
ITGB6
|
integrin subunit beta 6 |
| chr12_-_75209814 | 0.44 |
ENST00000549446.6
|
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr9_-_34589716 | 0.42 |
ENST00000378980.8
|
CNTFR
|
ciliary neurotrophic factor receptor |
| chr19_+_17795129 | 0.42 |
ENST00000600777.1
ENST00000318683.7 ENST00000595387.1 |
B3GNT3
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
| chr17_+_70075317 | 0.42 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr15_-_48878001 | 0.41 |
ENST00000558220.1
ENST00000537958.5 |
SHC4
|
SHC adaptor protein 4 |
| chr12_-_75209701 | 0.40 |
ENST00000350228.6
ENST00000298972.5 |
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr8_-_102238874 | 0.39 |
ENST00000519962.5
ENST00000519317.5 ENST00000395912.6 |
RRM2B
|
ribonucleotide reductase regulatory TP53 inducible subunit M2B |
| chr6_+_101393699 | 0.39 |
ENST00000369134.9
ENST00000684068.1 ENST00000683903.1 ENST00000681975.1 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr8_-_102238769 | 0.39 |
ENST00000621845.1
ENST00000522368.5 |
RRM2B
|
ribonucleotide reductase regulatory TP53 inducible subunit M2B |
| chr2_+_108377947 | 0.38 |
ENST00000272452.7
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr7_-_93226449 | 0.37 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr9_-_34589701 | 0.37 |
ENST00000351266.8
|
CNTFR
|
ciliary neurotrophic factor receptor |
| chr8_-_102238903 | 0.35 |
ENST00000251810.8
|
RRM2B
|
ribonucleotide reductase regulatory TP53 inducible subunit M2B |
| chr14_-_59630582 | 0.35 |
ENST00000395090.5
|
RTN1
|
reticulon 1 |
| chr19_+_41708635 | 0.35 |
ENST00000617332.4
ENST00000615021.4 ENST00000616453.1 ENST00000405816.5 ENST00000435837.2 |
CEACAM5
ENSG00000267881.2
|
CEA cell adhesion molecule 5 novel protein, readthrough between CEACAM5-CEACAM6 |
| chr2_-_206159410 | 0.34 |
ENST00000457011.5
ENST00000440274.5 ENST00000432169.5 ENST00000233190.11 |
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr1_-_11654422 | 0.34 |
ENST00000354287.5
|
FBXO2
|
F-box protein 2 |
| chr12_+_4269771 | 0.33 |
ENST00000676411.1
|
CCND2
|
cyclin D2 |
| chr5_+_141213919 | 0.33 |
ENST00000341948.6
|
PCDHB13
|
protocadherin beta 13 |
| chr6_-_46954922 | 0.31 |
ENST00000265417.7
|
ADGRF5
|
adhesion G protein-coupled receptor F5 |
| chr7_-_101245032 | 0.31 |
ENST00000442303.1
|
FIS1
|
fission, mitochondrial 1 |
| chr11_+_57181945 | 0.31 |
ENST00000497933.3
|
LRRC55
|
leucine rich repeat containing 55 |
| chr8_-_27611866 | 0.30 |
ENST00000519742.5
|
CLU
|
clusterin |
| chr5_+_151212117 | 0.28 |
ENST00000523466.5
|
GM2A
|
GM2 ganglioside activator |
| chrX_-_154354889 | 0.28 |
ENST00000438732.2
|
FLNA
|
filamin A |
| chr14_-_21580076 | 0.27 |
ENST00000641040.1
ENST00000641185.1 |
OR10G3
|
olfactory receptor family 10 subfamily G member 3 |
| chr3_+_48989876 | 0.27 |
ENST00000343546.8
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane |
| chr8_+_49911604 | 0.26 |
ENST00000642164.1
ENST00000644093.1 ENST00000643999.1 ENST00000647073.1 ENST00000646880.1 |
SNTG1
|
syntrophin gamma 1 |
| chr19_-_54364983 | 0.25 |
ENST00000434277.6
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1 |
| chr10_+_110207587 | 0.25 |
ENST00000332674.9
ENST00000453116.5 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr2_-_19901624 | 0.25 |
ENST00000431392.1
|
TTC32
|
tetratricopeptide repeat domain 32 |
| chr3_-_149576203 | 0.24 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr3_+_8501846 | 0.24 |
ENST00000454244.4
|
LMCD1
|
LIM and cysteine rich domains 1 |
| chr2_+_241869722 | 0.24 |
ENST00000343216.3
|
RTP5
|
receptor transporter protein 5 (putative) |
| chr7_-_101245056 | 0.24 |
ENST00000223136.5
|
FIS1
|
fission, mitochondrial 1 |
| chr4_-_106238690 | 0.23 |
ENST00000503516.1
|
TBCK
|
TBC1 domain containing kinase |
| chr3_+_69763726 | 0.22 |
ENST00000448226.9
|
MITF
|
melanocyte inducing transcription factor |
| chr5_-_78549151 | 0.22 |
ENST00000515007.6
|
LHFPL2
|
LHFPL tetraspan subfamily member 2 |
| chr13_+_45120493 | 0.22 |
ENST00000340473.8
|
GTF2F2
|
general transcription factor IIF subunit 2 |
| chr19_+_5914202 | 0.21 |
ENST00000588776.8
ENST00000588865.2 |
CAPS
|
calcyphosine |
| chr4_+_17577487 | 0.21 |
ENST00000606142.5
|
LAP3
|
leucine aminopeptidase 3 |
| chr14_-_106389858 | 0.21 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr3_+_25428233 | 0.21 |
ENST00000437042.6
ENST00000330688.9 |
RARB
|
retinoic acid receptor beta |
| chrX_-_45200895 | 0.21 |
ENST00000377934.4
|
DIPK2B
|
divergent protein kinase domain 2B |
| chr10_-_27243019 | 0.20 |
ENST00000679293.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr6_-_130970428 | 0.20 |
ENST00000529208.5
|
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr2_-_206159509 | 0.20 |
ENST00000423725.5
|
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr11_-_58844695 | 0.20 |
ENST00000287275.6
|
GLYATL2
|
glycine-N-acyltransferase like 2 |
| chr19_-_54364807 | 0.19 |
ENST00000474878.5
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1 |
| chr4_+_17577190 | 0.19 |
ENST00000226299.9
ENST00000618908.4 |
LAP3
|
leucine aminopeptidase 3 |
| chr1_-_246507237 | 0.19 |
ENST00000490107.6
|
SMYD3
|
SET and MYND domain containing 3 |
| chr6_+_4889992 | 0.18 |
ENST00000343762.5
|
CDYL
|
chromodomain Y like |
| chr18_-_27185284 | 0.18 |
ENST00000580774.2
ENST00000618847.5 |
CHST9
|
carbohydrate sulfotransferase 9 |
| chr7_+_148133684 | 0.18 |
ENST00000628930.2
|
CNTNAP2
|
contactin associated protein 2 |
| chr17_+_41967783 | 0.18 |
ENST00000585452.1
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr5_+_140882116 | 0.17 |
ENST00000289272.3
ENST00000409494.5 ENST00000617769.1 |
PCDHA13
|
protocadherin alpha 13 |
| chr11_-_62689523 | 0.17 |
ENST00000317449.5
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr12_-_54981838 | 0.16 |
ENST00000316577.12
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr15_+_90902218 | 0.15 |
ENST00000558290.5
ENST00000558853.5 ENST00000559999.5 |
MAN2A2
|
mannosidase alpha class 2A member 2 |
| chr14_+_99773698 | 0.15 |
ENST00000556199.1
|
EML1
|
EMAP like 1 |
| chr8_+_49911396 | 0.14 |
ENST00000642720.2
|
SNTG1
|
syntrophin gamma 1 |
| chr4_+_86936386 | 0.14 |
ENST00000511442.1
|
AFF1
|
AF4/FMR2 family member 1 |
| chr18_-_77127935 | 0.13 |
ENST00000581878.5
|
MBP
|
myelin basic protein |
| chr3_+_40505992 | 0.13 |
ENST00000420891.5
ENST00000314529.10 ENST00000418905.1 |
ZNF620
|
zinc finger protein 620 |
| chr8_-_51408974 | 0.13 |
ENST00000522933.5
|
PXDNL
|
peroxidasin like |
| chr8_-_98942557 | 0.12 |
ENST00000523601.5
|
STK3
|
serine/threonine kinase 3 |
| chr16_+_68539213 | 0.12 |
ENST00000570495.5
ENST00000564323.5 ENST00000562156.5 ENST00000573685.5 ENST00000563169.7 ENST00000611381.4 |
ZFP90
|
ZFP90 zinc finger protein |
| chr16_-_29505820 | 0.11 |
ENST00000550665.5
|
NPIPB12
|
nuclear pore complex interacting protein family member B12 |
| chr14_-_102240597 | 0.11 |
ENST00000523231.5
ENST00000524370.5 ENST00000517966.5 |
MOK
|
MOK protein kinase |
| chr14_+_20138820 | 0.10 |
ENST00000641086.1
|
OR4N5
|
olfactory receptor family 4 subfamily N member 5 |
| chr11_+_8911280 | 0.10 |
ENST00000530281.5
ENST00000396648.6 ENST00000534147.5 ENST00000529942.1 |
AKIP1
|
A-kinase interacting protein 1 |
| chr19_-_54364863 | 0.10 |
ENST00000348231.8
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1 |
| chr17_-_74592283 | 0.10 |
ENST00000375352.1
|
CD300LD
|
CD300 molecule like family member d |
| chr14_-_92121902 | 0.09 |
ENST00000329559.8
|
NDUFB1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chrX_-_70931089 | 0.09 |
ENST00000374299.8
ENST00000298085.4 |
SLC7A3
|
solute carrier family 7 member 3 |
| chr6_-_31958431 | 0.09 |
ENST00000625905.1
ENST00000454913.5 ENST00000436289.6 |
NELFE
|
negative elongation factor complex member E |
| chr14_+_100381974 | 0.09 |
ENST00000542471.2
|
WDR25
|
WD repeat domain 25 |
| chr4_+_154743993 | 0.08 |
ENST00000336356.4
|
LRAT
|
lecithin retinol acyltransferase |
| chr10_+_70052806 | 0.08 |
ENST00000676609.1
ENST00000679349.1 ENST00000373255.9 ENST00000676608.1 ENST00000455786.1 |
MACROH2A2
|
macroH2A.2 histone |
| chr20_-_10431922 | 0.08 |
ENST00000399054.6
ENST00000609375.1 |
MKKS
ENSG00000285508.1
|
McKusick-Kaufman syndrome novel protein |
| chr17_+_42798857 | 0.08 |
ENST00000588408.6
|
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr7_-_100100716 | 0.08 |
ENST00000354230.7
ENST00000425308.5 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr7_+_1086800 | 0.08 |
ENST00000413368.5
ENST00000397092.5 ENST00000297469.3 |
GPER1
|
G protein-coupled estrogen receptor 1 |
| chr7_+_128758947 | 0.08 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr2_+_237968770 | 0.08 |
ENST00000434655.5
ENST00000612130.4 |
UBE2F
|
ubiquitin conjugating enzyme E2 F (putative) |
| chr1_-_85404494 | 0.08 |
ENST00000633113.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr10_-_60139733 | 0.08 |
ENST00000506635.5
|
ANK3
|
ankyrin 3 |
| chr9_-_77648303 | 0.07 |
ENST00000341700.7
|
GNA14
|
G protein subunit alpha 14 |
| chr19_+_17309531 | 0.07 |
ENST00000359866.9
|
DDA1
|
DET1 and DDB1 associated 1 |
| chr19_+_17794776 | 0.07 |
ENST00000599265.5
|
B3GNT3
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
| chr9_-_6015607 | 0.06 |
ENST00000485372.1
ENST00000259569.6 ENST00000623170.1 |
RANBP6
|
RAN binding protein 6 |
| chr7_-_92528446 | 0.06 |
ENST00000248633.9
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr7_-_92528433 | 0.06 |
ENST00000428214.5
ENST00000438045.5 |
PEX1
|
peroxisomal biogenesis factor 1 |
| chr12_-_54973547 | 0.05 |
ENST00000526532.5
ENST00000532757.5 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr10_+_97849824 | 0.05 |
ENST00000370602.6
|
GOLGA7B
|
golgin A7 family member B |
| chr1_-_31919368 | 0.05 |
ENST00000457805.6
ENST00000602725.5 ENST00000679970.1 |
PTP4A2
ENSG00000288678.1
|
protein tyrosine phosphatase 4A2 novel protein |
| chr19_-_43596123 | 0.05 |
ENST00000422989.6
ENST00000598324.1 |
IRGQ
|
immunity related GTPase Q |
| chr17_+_47651061 | 0.05 |
ENST00000540627.5
|
KPNB1
|
karyopherin subunit beta 1 |
| chr19_-_46784905 | 0.04 |
ENST00000594991.5
|
SLC1A5
|
solute carrier family 1 member 5 |
| chr17_+_6755559 | 0.04 |
ENST00000574394.5
|
XAF1
|
XIAP associated factor 1 |
| chr3_+_185282941 | 0.04 |
ENST00000448876.5
ENST00000446828.5 ENST00000447637.1 ENST00000424227.5 ENST00000454237.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr3_+_63819368 | 0.04 |
ENST00000616659.1
|
C3orf49
|
chromosome 3 open reading frame 49 |
| chr9_+_133376334 | 0.04 |
ENST00000371957.4
|
STKLD1
|
serine/threonine kinase like domain containing 1 |
| chr2_-_55296361 | 0.03 |
ENST00000647547.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr22_+_37849437 | 0.03 |
ENST00000406934.5
ENST00000451427.1 |
EIF3L
|
eukaryotic translation initiation factor 3 subunit L |
| chr12_-_11062294 | 0.03 |
ENST00000533467.1
|
TAS2R46
|
taste 2 receptor member 46 |
| chr17_+_27471999 | 0.03 |
ENST00000583370.5
ENST00000509603.6 ENST00000268763.10 ENST00000398988.7 |
KSR1
|
kinase suppressor of ras 1 |
| chr1_+_173824956 | 0.03 |
ENST00000648458.1
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr17_-_42798680 | 0.03 |
ENST00000328434.8
|
COA3
|
cytochrome c oxidase assembly factor 3 |
| chr10_+_79841358 | 0.03 |
ENST00000602967.5
|
NUTM2E
|
NUT family member 2E |
| chr16_+_33009175 | 0.03 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr2_-_222298974 | 0.03 |
ENST00000392069.6
ENST00000392070.7 |
PAX3
|
paired box 3 |
| chr20_-_23751256 | 0.02 |
ENST00000398402.1
|
CST1
|
cystatin SN |
| chr13_+_101489940 | 0.01 |
ENST00000376162.7
|
ITGBL1
|
integrin subunit beta like 1 |
| chr1_-_246507312 | 0.01 |
ENST00000403792.7
|
SMYD3
|
SET and MYND domain containing 3 |
| chr1_-_243254460 | 0.01 |
ENST00000522191.5
|
CEP170
|
centrosomal protein 170 |
| chr3_+_122680850 | 0.01 |
ENST00000494811.2
|
PARP14
|
poly(ADP-ribose) polymerase family member 14 |
| chr2_+_74549026 | 0.01 |
ENST00000409429.5
|
DOK1
|
docking protein 1 |
| chr14_+_51847145 | 0.01 |
ENST00000615906.4
|
GNG2
|
G protein subunit gamma 2 |
| chr1_-_248682328 | 0.00 |
ENST00000342623.5
|
OR14I1
|
olfactory receptor family 14 subfamily I member 1 |
| chr15_+_43593601 | 0.00 |
ENST00000449946.5
ENST00000417289.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr22_+_37849372 | 0.00 |
ENST00000624234.3
ENST00000381683.10 ENST00000652021.1 ENST00000414316.5 |
EIF3L
|
eukaryotic translation initiation factor 3 subunit L |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFAT5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.3 | 1.6 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.3 | 1.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 0.7 | GO:0002434 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) response to cyclosporin A(GO:1905237) |
| 0.2 | 2.8 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.2 | 0.8 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.2 | 1.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.7 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 2.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 2.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 0.5 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.1 | 2.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.4 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 1.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.7 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 0.5 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.8 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.1 | 0.6 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.3 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 1.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 1.7 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.8 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.8 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 3.0 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.3 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.6 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.3 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 0.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 1.1 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.3 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.5 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 0.0 | 0.2 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.7 | GO:0015991 | vacuolar acidification(GO:0007035) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 3.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.8 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.4 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 1.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.9 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.0 | GO:1903803 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.3 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.0 | GO:0070145 | mitochondrial asparaginyl-tRNA aminoacylation(GO:0070145) |
| 0.0 | 0.8 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.3 | 0.8 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.2 | 3.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 1.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 0.8 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 0.5 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 1.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 2.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.3 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 2.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.7 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 1.6 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 1.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 3.8 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.8 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 3.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.2 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 1.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.2 | 0.7 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 0.7 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.2 | 1.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.5 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.1 | 1.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.8 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.9 | GO:0008443 | phosphofructokinase activity(GO:0008443) |
| 0.1 | 1.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.3 | GO:0032428 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.1 | 3.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 2.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.8 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 0.6 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.0 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 1.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.7 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 2.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.0 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) aspartate-tRNA(Asn) ligase activity(GO:0050560) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 3.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 1.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |