Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
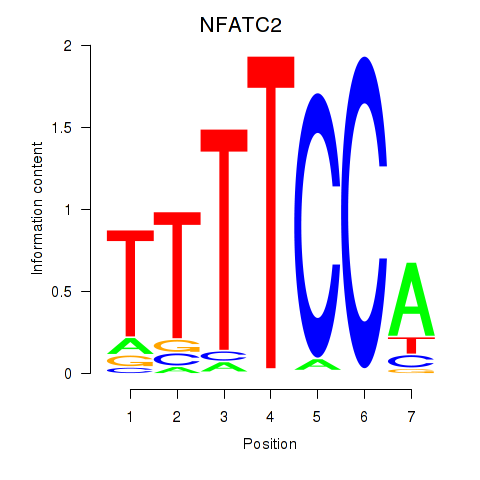
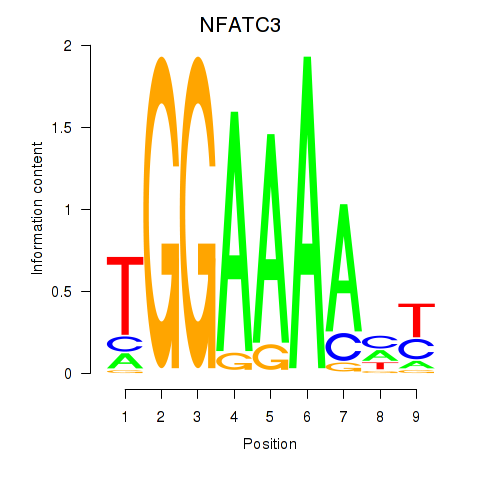
Results for NFATC2_NFATC3
Z-value: 1.61
Transcription factors associated with NFATC2_NFATC3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC2
|
ENSG00000101096.20 | nuclear factor of activated T cells 2 |
|
NFATC3
|
ENSG00000072736.19 | nuclear factor of activated T cells 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC2 | hg38_v1_chr20_-_51562829_51562853 | -0.54 | 1.3e-03 | Click! |
| NFATC3 | hg38_v1_chr16_+_68085861_68085968 | -0.29 | 1.0e-01 | Click! |
Activity profile of NFATC2_NFATC3 motif
Sorted Z-values of NFATC2_NFATC3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC2_NFATC3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.6 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 1.6 | 6.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.5 | 4.4 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 1.4 | 18.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 1.4 | 5.6 | GO:0009956 | radial pattern formation(GO:0009956) |
| 1.3 | 6.7 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 1.3 | 24.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.2 | 5.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 1.2 | 7.2 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 1.1 | 4.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.9 | 2.8 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.9 | 3.5 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.9 | 3.4 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.8 | 5.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.8 | 1.7 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.8 | 2.4 | GO:0060721 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.7 | 5.9 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.7 | 3.5 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.7 | 2.8 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.7 | 2.8 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.7 | 2.0 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.7 | 4.8 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.7 | 2.7 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.7 | 2.0 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.6 | 7.8 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.6 | 6.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.6 | 5.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.6 | 1.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.6 | 1.8 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.6 | 6.1 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.5 | 3.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.5 | 1.5 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.5 | 1.5 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.5 | 9.7 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.5 | 1.4 | GO:1990051 | negative regulation of glucagon secretion(GO:0070093) activation of protein kinase C activity(GO:1990051) regulation of glutamine transport(GO:2000485) |
| 0.5 | 1.4 | GO:0036323 | vascular endothelial growth factor receptor-1 signaling pathway(GO:0036323) |
| 0.4 | 1.8 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.4 | 5.0 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.4 | 1.2 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 0.4 | 1.2 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.4 | 3.3 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.4 | 3.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.4 | 1.6 | GO:0061443 | endocardial cushion cell differentiation(GO:0061443) |
| 0.4 | 4.3 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.4 | 3.5 | GO:0072343 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.4 | 1.9 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.4 | 0.8 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.4 | 3.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.4 | 1.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.4 | 4.6 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.4 | 5.7 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.4 | 2.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 | 2.8 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.3 | 2.4 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.3 | 1.7 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.3 | 1.0 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.3 | 0.3 | GO:0097102 | endothelial tip cell fate specification(GO:0097102) |
| 0.3 | 1.3 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.3 | 2.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.3 | 0.9 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.3 | 0.9 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.3 | 1.2 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.3 | 5.2 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.3 | 1.4 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.3 | 1.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.3 | 1.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.3 | 1.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.3 | 1.8 | GO:0032439 | endosome localization(GO:0032439) |
| 0.3 | 4.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 1.5 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 1.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 3.2 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.2 | 3.4 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.2 | 7.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 1.0 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) response to prolactin(GO:1990637) |
| 0.2 | 11.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 0.7 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.2 | 0.7 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.2 | 2.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 4.4 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.2 | 2.6 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.2 | 0.9 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 1.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.2 | 2.8 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.2 | 0.4 | GO:0003274 | endocardial cushion fusion(GO:0003274) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.2 | 1.8 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.2 | 2.6 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 1.0 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.2 | 3.3 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.2 | 4.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 | 1.5 | GO:0035931 | mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.2 | 1.8 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.2 | 0.9 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.2 | 0.9 | GO:0035645 | enteric smooth muscle cell differentiation(GO:0035645) |
| 0.2 | 1.0 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 0.5 | GO:0061027 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) |
| 0.2 | 8.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.2 | 1.2 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.2 | 1.8 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 0.5 | GO:1904528 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) positive regulation of microtubule binding(GO:1904528) |
| 0.2 | 2.4 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.2 | 0.5 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.1 | 1.6 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.7 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 2.7 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 | 0.4 | GO:0021758 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.1 | 3.2 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 3.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 4.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.7 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 1.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.5 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.5 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 3.9 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 2.0 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.6 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.5 | GO:0014806 | smooth muscle hyperplasia(GO:0014806) |
| 0.1 | 2.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.4 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.1 | 1.5 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.1 | 1.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.3 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 0.5 | GO:0051946 | lactate biosynthetic process(GO:0019249) regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 | 1.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 1.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.3 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.1 | 0.7 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.3 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.3 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.1 | 0.5 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 1.4 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 1.9 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.5 | GO:0034239 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 1.9 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 1.8 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 1.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.8 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.3 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.4 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 0.4 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 9.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.4 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.8 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 1.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 2.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.8 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 1.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.3 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.8 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.4 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.1 | 1.5 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 2.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.3 | GO:0061551 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) trigeminal ganglion development(GO:0061551) neural crest cell migration involved in sympathetic nervous system development(GO:1903045) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 0.6 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.5 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.2 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 0.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 2.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.7 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 1.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 2.3 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 2.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 1.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 1.9 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 15.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.5 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.3 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.1 | 0.4 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.1 | 1.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 1.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 2.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.3 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.1 | 0.8 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 1.3 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 0.7 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.4 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.1 | 0.5 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 1.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 1.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.3 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.2 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.4 | GO:0046086 | AMP catabolic process(GO:0006196) adenosine biosynthetic process(GO:0046086) |
| 0.1 | 1.5 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 0.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) dehydroascorbic acid transport(GO:0070837) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 0.5 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.1 | 1.7 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.1 | 0.4 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 1.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.9 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 1.6 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.4 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 3.2 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 1.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 7.3 | GO:2000257 | regulation of complement activation(GO:0030449) regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.1 | GO:0072249 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 2.1 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.5 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 2.0 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.9 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.8 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 1.2 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.3 | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.0 | 0.7 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 1.0 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.3 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.8 | GO:0060749 | mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.0 | 0.1 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.0 | 0.4 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 1.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.7 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.4 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.4 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 4.6 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.5 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 1.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.2 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 1.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.4 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 1.8 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 1.5 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.0 | 0.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 3.2 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 0.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 2.9 | GO:0045931 | positive regulation of mitotic cell cycle(GO:0045931) |
| 0.0 | 1.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 2.3 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.1 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 2.9 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.2 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.0 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.0 | 0.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.9 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.6 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.7 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.4 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.4 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.4 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.3 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 4.7 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.4 | GO:0090109 | regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) regulation of adherens junction organization(GO:1903391) |
| 0.0 | 0.1 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 1.0 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.1 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.6 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 31.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 2.0 | 5.9 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 1.9 | 18.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.1 | 5.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 1.0 | 4.8 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 0.8 | 2.4 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.7 | 2.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.6 | 4.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.5 | 19.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.5 | 1.5 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.5 | 1.9 | GO:0071757 | hexameric IgM immunoglobulin complex(GO:0071757) |
| 0.4 | 7.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.4 | 6.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.4 | 1.9 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 0.4 | 4.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.3 | 5.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 2.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 1.5 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.3 | 3.2 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.3 | 0.8 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 2.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 6.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 2.7 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 2.5 | GO:0098984 | neuron to neuron synapse(GO:0098984) |
| 0.2 | 1.3 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.2 | 2.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.2 | 4.7 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.2 | 1.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 4.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 0.7 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 1.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 4.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 0.5 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 5.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.9 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 1.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 2.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 1.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 2.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.6 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 1.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 9.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 4.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 3.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.2 | GO:0044207 | eukaryotic 48S preinitiation complex(GO:0033290) translation initiation ternary complex(GO:0044207) |
| 0.1 | 9.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 2.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 17.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 5.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 2.3 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 7.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.9 | GO:1990752 | microtubule end(GO:1990752) |
| 0.1 | 0.9 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 1.8 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.4 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 2.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.4 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 1.2 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 1.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 2.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 4.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.4 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 1.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 2.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 8.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 3.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 15.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 4.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 9.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 1.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 1.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 1.0 | 5.9 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.9 | 2.6 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.8 | 6.6 | GO:0043120 | interleukin-8 binding(GO:0019959) tumor necrosis factor binding(GO:0043120) |
| 0.8 | 3.3 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.7 | 3.7 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.7 | 5.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.7 | 2.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.6 | 5.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.5 | 3.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.5 | 1.4 | GO:0036326 | VEGF-A-activated receptor activity(GO:0036326) VEGF-B-activated receptor activity(GO:0036327) placental growth factor-activated receptor activity(GO:0036332) |
| 0.4 | 1.9 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.4 | 3.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.4 | 2.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 16.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.3 | 3.8 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.3 | 1.7 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.3 | 2.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.3 | 3.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 1.9 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.3 | 7.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 46.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.3 | 2.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 3.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.3 | 4.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 1.9 | GO:0019862 | IgA binding(GO:0019862) |
| 0.3 | 0.8 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.3 | 5.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 1.4 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.2 | 1.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 0.7 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.2 | 7.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 0.9 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.2 | 0.9 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.2 | 1.3 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.2 | 2.4 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.2 | 1.5 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.2 | 7.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 1.4 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.2 | 1.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 1.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 1.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 3.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 1.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 7.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 7.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 1.8 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.2 | 4.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 2.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 5.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.8 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 1.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 3.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 2.7 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.2 | 0.6 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 1.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.4 | GO:0070975 | FHA domain binding(GO:0070975) |
| 0.1 | 0.4 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 1.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 2.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.8 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.6 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.1 | 0.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.5 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 1.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.4 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 0.4 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 1.4 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 2.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.9 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 1.0 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.9 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 1.5 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.7 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 1.5 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 1.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.8 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.9 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.1 | 4.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 18.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.7 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 1.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.7 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 5.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.5 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.5 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 1.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.6 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.8 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 0.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 8.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 2.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.3 | GO:0048030 | lactose binding(GO:0030395) disaccharide binding(GO:0048030) |
| 0.1 | 2.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.0 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 5.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 4.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 2.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.6 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 1.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.2 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 1.3 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 1.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.4 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.4 | GO:0044020 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 1.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 1.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.5 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.2 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.9 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.5 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 1.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.3 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 1.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 19.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.2 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 1.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 12.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 5.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.4 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 1.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 4.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 2.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.3 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 2.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 1.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 34.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 16.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 10.7 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 3.5 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 2.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 41.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 6.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 2.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 0.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 6.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 5.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 11.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 23.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 5.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 5.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 15.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.4 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 2.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.7 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 22.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.6 | 9.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 9.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 8.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 4.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 23.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 9.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 3.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 9.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 7.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.9 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.1 | 4.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 4.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 14.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 3.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.0 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 1.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 3.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 3.8 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 2.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 4.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 2.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 5.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.9 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.9 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 2.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.0 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 1.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |