Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
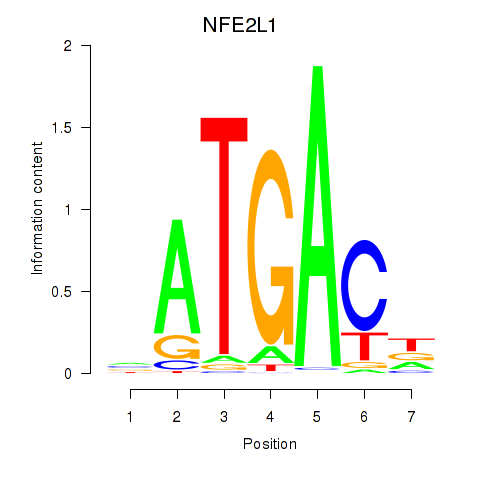
Results for NFE2L1
Z-value: 0.94
Transcription factors associated with NFE2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFE2L1
|
ENSG00000082641.16 | nuclear factor, erythroid 2 like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFE2L1 | hg38_v1_chr17_+_48055945_48055986 | -0.57 | 6.6e-04 | Click! |
Activity profile of NFE2L1 motif
Sorted Z-values of NFE2L1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_59019397 | 3.73 |
ENST00000217133.2
|
TUBB1
|
tubulin beta 1 class VI |
| chr1_+_192158448 | 3.07 |
ENST00000367460.4
|
RGS18
|
regulator of G protein signaling 18 |
| chr2_-_157488829 | 2.71 |
ENST00000435117.1
ENST00000439355.5 |
CYTIP
|
cytohesin 1 interacting protein |
| chr7_+_142352802 | 2.66 |
ENST00000634605.1
|
TRBV7-2
|
T cell receptor beta variable 7-2 |
| chr7_+_142469521 | 2.61 |
ENST00000390371.3
|
TRBV6-6
|
T cell receptor beta variable 6-6 |
| chr4_+_73740541 | 2.39 |
ENST00000401931.1
ENST00000307407.8 |
CXCL8
|
C-X-C motif chemokine ligand 8 |
| chr1_-_183569186 | 2.34 |
ENST00000420553.5
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr12_-_122716790 | 2.23 |
ENST00000528880.3
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr3_-_27722699 | 2.06 |
ENST00000461503.2
|
EOMES
|
eomesodermin |
| chr12_-_14951106 | 2.02 |
ENST00000541644.5
ENST00000545895.5 |
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr17_-_35880350 | 1.99 |
ENST00000605140.6
ENST00000651122.1 ENST00000603197.6 |
CCL5
|
C-C motif chemokine ligand 5 |
| chr12_-_14950606 | 1.96 |
ENST00000536592.5
|
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr12_-_122703346 | 1.86 |
ENST00000328880.6
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr7_+_80638633 | 1.82 |
ENST00000447544.7
ENST00000482059.6 |
CD36
|
CD36 molecule |
| chr4_-_154590735 | 1.69 |
ENST00000403106.8
ENST00000622532.1 ENST00000651975.1 |
FGA
|
fibrinogen alpha chain |
| chr16_+_72056153 | 1.65 |
ENST00000576168.6
ENST00000567185.7 ENST00000567612.2 |
HP
|
haptoglobin |
| chr11_+_60455839 | 1.65 |
ENST00000532491.5
ENST00000532073.5 ENST00000345732.9 ENST00000534668.6 ENST00000528313.1 ENST00000533306.6 ENST00000674194.1 |
MS4A1
|
membrane spanning 4-domains A1 |
| chr7_+_80638662 | 1.61 |
ENST00000394788.7
|
CD36
|
CD36 molecule |
| chr7_+_80646436 | 1.60 |
ENST00000419819.2
|
CD36
|
CD36 molecule |
| chrX_+_124346544 | 1.55 |
ENST00000371139.9
|
SH2D1A
|
SH2 domain containing 1A |
| chr12_-_52493250 | 1.54 |
ENST00000330722.7
|
KRT6A
|
keratin 6A |
| chr9_-_114930508 | 1.53 |
ENST00000223795.3
ENST00000618336.4 |
TNFSF8
|
TNF superfamily member 8 |
| chr14_+_22147988 | 1.52 |
ENST00000390457.2
|
TRAV27
|
T cell receptor alpha variable 27 |
| chr7_+_80646347 | 1.49 |
ENST00000413265.5
|
CD36
|
CD36 molecule |
| chr7_+_142618821 | 1.46 |
ENST00000390393.3
|
TRBV19
|
T cell receptor beta variable 19 |
| chr14_+_21887848 | 1.45 |
ENST00000390437.2
|
TRAV12-2
|
T cell receptor alpha variable 12-2 |
| chr6_-_132734692 | 1.44 |
ENST00000509351.5
ENST00000417437.6 ENST00000423615.6 ENST00000427187.6 ENST00000414302.7 ENST00000367927.9 ENST00000450865.2 |
VNN3
|
vanin 3 |
| chr19_-_54100792 | 1.43 |
ENST00000391761.5
ENST00000356532.7 ENST00000616447.4 ENST00000359649.8 ENST00000358375.8 ENST00000391760.1 ENST00000351806.8 |
OSCAR
|
osteoclast associated Ig-like receptor |
| chr16_+_31355215 | 1.39 |
ENST00000562522.2
|
ITGAX
|
integrin subunit alpha X |
| chr6_+_144150492 | 1.38 |
ENST00000367568.5
|
STX11
|
syntaxin 11 |
| chr14_+_22462932 | 1.36 |
ENST00000390477.2
|
TRDC
|
T cell receptor delta constant |
| chr16_+_31355165 | 1.35 |
ENST00000562918.5
ENST00000268296.9 |
ITGAX
|
integrin subunit alpha X |
| chr5_+_169695135 | 1.35 |
ENST00000523684.5
ENST00000519734.5 |
DOCK2
|
dedicator of cytokinesis 2 |
| chr7_+_142313144 | 1.34 |
ENST00000390357.3
|
TRBV4-1
|
T cell receptor beta variable 4-1 |
| chrX_+_124346525 | 1.33 |
ENST00000360027.4
|
SH2D1A
|
SH2 domain containing 1A |
| chr10_+_69088096 | 1.30 |
ENST00000242465.4
|
SRGN
|
serglycin |
| chr7_+_80646305 | 1.28 |
ENST00000426978.5
ENST00000432207.5 |
CD36
|
CD36 molecule |
| chr18_+_3252208 | 1.27 |
ENST00000578562.6
|
MYL12A
|
myosin light chain 12A |
| chr2_-_196176467 | 1.27 |
ENST00000409228.5
|
STK17B
|
serine/threonine kinase 17b |
| chr20_-_36646146 | 1.27 |
ENST00000262866.9
|
SLA2
|
Src like adaptor 2 |
| chr6_+_158649997 | 1.26 |
ENST00000360448.8
ENST00000367081.7 ENST00000611299.5 |
SYTL3
|
synaptotagmin like 3 |
| chr14_-_22815421 | 1.26 |
ENST00000674313.1
ENST00000555959.1 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr5_-_177509814 | 1.26 |
ENST00000510898.7
ENST00000502885.5 ENST00000506493.5 |
DOK3
|
docking protein 3 |
| chr13_-_99307387 | 1.24 |
ENST00000376414.5
|
GPR183
|
G protein-coupled receptor 183 |
| chr3_-_115071333 | 1.20 |
ENST00000462705.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr2_+_169069537 | 1.19 |
ENST00000428522.5
ENST00000450153.1 ENST00000674881.1 ENST00000421653.5 |
DHRS9
|
dehydrogenase/reductase 9 |
| chr11_+_35176696 | 1.18 |
ENST00000528455.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr2_-_196176320 | 1.18 |
ENST00000420683.1
|
STK17B
|
serine/threonine kinase 17b |
| chr18_+_3252267 | 1.15 |
ENST00000536605.1
ENST00000580887.5 |
MYL12A
|
myosin light chain 12A |
| chr19_-_54242751 | 1.15 |
ENST00000245621.6
ENST00000396365.6 |
LILRA6
|
leukocyte immunoglobulin like receptor A6 |
| chr4_+_73853290 | 1.14 |
ENST00000226524.4
|
PF4V1
|
platelet factor 4 variant 1 |
| chr17_-_66220630 | 1.13 |
ENST00000585162.1
|
APOH
|
apolipoprotein H |
| chr12_-_9999176 | 1.13 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr1_+_198638457 | 1.12 |
ENST00000367379.6
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr12_-_54419259 | 1.11 |
ENST00000293379.9
|
ITGA5
|
integrin subunit alpha 5 |
| chr11_+_35176639 | 1.11 |
ENST00000527889.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr15_+_88639009 | 1.10 |
ENST00000306072.10
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr11_+_35180342 | 1.10 |
ENST00000639002.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr14_+_21997531 | 1.10 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr11_+_35176575 | 1.07 |
ENST00000526000.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr19_-_43656616 | 1.07 |
ENST00000593447.5
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr7_+_142424942 | 1.05 |
ENST00000426318.2
|
TRBV10-2
|
T cell receptor beta variable 10-2 |
| chr7_+_142615710 | 1.05 |
ENST00000611520.1
|
TRBV18
|
T cell receptor beta variable 18 |
| chr21_-_14546297 | 1.04 |
ENST00000400566.6
ENST00000400564.5 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr11_+_35189869 | 1.04 |
ENST00000525688.5
ENST00000278385.10 ENST00000533222.5 |
CD44
|
CD44 molecule (Indian blood group) |
| chr1_+_198638968 | 1.02 |
ENST00000348564.11
ENST00000530727.5 ENST00000442510.8 ENST00000645247.1 ENST00000367367.8 ENST00000367364.5 ENST00000413409.6 |
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr19_-_42217667 | 1.01 |
ENST00000336034.8
ENST00000596251.6 ENST00000598200.1 ENST00000598727.5 |
DEDD2
|
death effector domain containing 2 |
| chr11_+_35176611 | 1.01 |
ENST00000279452.10
|
CD44
|
CD44 molecule (Indian blood group) |
| chr15_-_60391127 | 1.01 |
ENST00000559350.5
ENST00000558986.5 ENST00000560389.5 |
ANXA2
|
annexin A2 |
| chr1_-_173917281 | 1.01 |
ENST00000367698.4
|
SERPINC1
|
serpin family C member 1 |
| chr4_-_15681454 | 1.00 |
ENST00000507899.1
ENST00000510802.5 |
FBXL5
|
F-box and leucine rich repeat protein 5 |
| chr14_+_51847116 | 1.00 |
ENST00000553560.5
|
GNG2
|
G protein subunit gamma 2 |
| chr9_-_125484490 | 1.00 |
ENST00000444226.1
|
MAPKAP1
|
MAPK associated protein 1 |
| chr14_+_22271921 | 0.99 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr14_+_22070548 | 0.98 |
ENST00000390450.3
|
TRAV22
|
T cell receptor alpha variable 22 |
| chr5_-_177509843 | 0.98 |
ENST00000510380.5
ENST00000357198.9 |
DOK3
|
docking protein 3 |
| chr2_+_218400039 | 0.98 |
ENST00000452977.5
|
CTDSP1
|
CTD small phosphatase 1 |
| chr2_+_201129483 | 0.97 |
ENST00000440180.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr14_+_22226711 | 0.96 |
ENST00000390463.3
|
TRAV36DV7
|
T cell receptor alpha variable 36/delta variable 7 |
| chr17_-_31318818 | 0.95 |
ENST00000578584.5
|
ENSG00000265118.5
|
novel protein |
| chr1_+_241532121 | 0.93 |
ENST00000366558.7
|
KMO
|
kynurenine 3-monooxygenase |
| chr2_+_169066994 | 0.92 |
ENST00000357546.6
ENST00000432060.6 |
DHRS9
|
dehydrogenase/reductase 9 |
| chr18_-_69947415 | 0.92 |
ENST00000577287.5
|
CD226
|
CD226 molecule |
| chr7_+_142626642 | 0.92 |
ENST00000390394.3
|
TRBV20-1
|
T cell receptor beta variable 20-1 |
| chr14_+_20781139 | 0.91 |
ENST00000304677.3
|
RNASE6
|
ribonuclease A family member k6 |
| chr15_+_88638947 | 0.91 |
ENST00000559876.2
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr1_+_198639162 | 0.91 |
ENST00000418674.1
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr6_-_2841853 | 0.90 |
ENST00000380739.6
|
SERPINB1
|
serpin family B member 1 |
| chr11_+_59713403 | 0.90 |
ENST00000641815.1
|
STX3
|
syntaxin 3 |
| chr2_+_90172802 | 0.90 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr1_+_28914614 | 0.89 |
ENST00000645184.1
|
EPB41
|
erythrocyte membrane protein band 4.1 |
| chr16_+_31033092 | 0.89 |
ENST00000394998.5
|
STX4
|
syntaxin 4 |
| chr7_+_142544199 | 0.89 |
ENST00000611462.1
|
TRBV10-3
|
T cell receptor beta variable 10-3 |
| chr19_-_54063905 | 0.89 |
ENST00000645936.1
ENST00000376626.5 ENST00000366170.6 ENST00000425006.3 |
VSTM1
|
V-set and transmembrane domain containing 1 |
| chr1_-_206923183 | 0.89 |
ENST00000525793.5
|
FCMR
|
Fc fragment of IgM receptor |
| chr17_+_59707636 | 0.89 |
ENST00000262291.9
ENST00000587945.1 ENST00000589823.6 ENST00000592106.5 ENST00000591315.5 |
VMP1
|
vacuole membrane protein 1 |
| chr16_+_28985043 | 0.88 |
ENST00000395456.7
ENST00000564277.5 ENST00000630764.2 ENST00000354453.7 |
LAT
|
linker for activation of T cells |
| chr1_+_28914597 | 0.88 |
ENST00000349460.9
|
EPB41
|
erythrocyte membrane protein band 4.1 |
| chr2_+_218245426 | 0.88 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex subunit 2 |
| chr17_+_44004604 | 0.87 |
ENST00000293404.8
ENST00000589767.1 |
NAGS
|
N-acetylglutamate synthase |
| chr7_+_142592928 | 0.86 |
ENST00000616518.1
|
TRBV15
|
T cell receptor beta variable 15 |
| chr6_+_41053194 | 0.85 |
ENST00000244669.3
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme catalytic subunit 2 |
| chr17_+_82237134 | 0.85 |
ENST00000583025.1
|
SLC16A3
|
solute carrier family 16 member 3 |
| chr7_+_142500027 | 0.85 |
ENST00000390375.2
|
TRBV5-6
|
T cell receptor beta variable 5-6 |
| chr11_+_118883884 | 0.83 |
ENST00000292174.5
|
CXCR5
|
C-X-C motif chemokine receptor 5 |
| chr2_+_201129318 | 0.83 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr14_+_21712313 | 0.83 |
ENST00000390424.2
|
TRAV2
|
T cell receptor alpha variable 2 |
| chr11_-_126268810 | 0.81 |
ENST00000332118.11
|
SRPRA
|
SRP receptor subunit alpha |
| chr6_-_32816910 | 0.80 |
ENST00000447394.1
ENST00000438763.7 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr2_+_203936755 | 0.80 |
ENST00000316386.11
ENST00000435193.1 |
ICOS
|
inducible T cell costimulator |
| chr7_+_142580911 | 0.79 |
ENST00000621184.1
|
TRBV12-5
|
T cell receptor beta variable 12-5 |
| chr5_-_180815563 | 0.79 |
ENST00000513431.5
ENST00000514438.1 |
MGAT1
|
alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase |
| chr9_-_114387973 | 0.79 |
ENST00000374088.8
|
AKNA
|
AT-hook transcription factor |
| chr1_-_92486049 | 0.79 |
ENST00000427103.5
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr19_-_43656540 | 0.78 |
ENST00000595038.5
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr12_-_10409757 | 0.78 |
ENST00000309384.2
|
KLRC4
|
killer cell lectin like receptor C4 |
| chr3_-_120647018 | 0.77 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr19_+_41003946 | 0.77 |
ENST00000593831.1
|
CYP2B6
|
cytochrome P450 family 2 subfamily B member 6 |
| chr13_-_99258366 | 0.76 |
ENST00000397470.5
ENST00000397473.7 |
GPR18
|
G protein-coupled receptor 18 |
| chr22_+_22322452 | 0.76 |
ENST00000390290.3
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr8_+_23528995 | 0.75 |
ENST00000523930.1
|
SLC25A37
|
solute carrier family 25 member 37 |
| chr6_-_55875583 | 0.75 |
ENST00000370830.4
|
BMP5
|
bone morphogenetic protein 5 |
| chr4_-_40630588 | 0.75 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr7_-_115968302 | 0.74 |
ENST00000457268.5
|
TFEC
|
transcription factor EC |
| chr14_-_22815493 | 0.74 |
ENST00000555911.1
|
SLC7A7
|
solute carrier family 7 member 7 |
| chrX_-_78327626 | 0.74 |
ENST00000614798.1
|
CYSLTR1
|
cysteinyl leukotriene receptor 1 |
| chr14_+_21978440 | 0.74 |
ENST00000390443.3
|
TRAV8-6
|
T cell receptor alpha variable 8-6 |
| chr14_+_21918161 | 0.73 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2 |
| chrX_+_124346571 | 0.73 |
ENST00000477673.2
|
SH2D1A
|
SH2 domain containing 1A |
| chr1_+_171090892 | 0.73 |
ENST00000367755.9
ENST00000479749.1 |
FMO3
|
flavin containing dimethylaniline monoxygenase 3 |
| chr2_-_210315160 | 0.73 |
ENST00000352451.4
|
MYL1
|
myosin light chain 1 |
| chr1_-_146021724 | 0.73 |
ENST00000475797.1
ENST00000497365.5 ENST00000336751.11 ENST00000634927.1 ENST00000421822.2 |
HJV
|
hemojuvelin BMP co-receptor |
| chr15_+_81296913 | 0.72 |
ENST00000394652.6
|
IL16
|
interleukin 16 |
| chr16_+_21612637 | 0.72 |
ENST00000568826.1
|
METTL9
|
methyltransferase like 9 |
| chr17_-_7252456 | 0.72 |
ENST00000571409.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr9_-_92482461 | 0.71 |
ENST00000651738.1
|
ASPN
|
asporin |
| chr21_+_29300111 | 0.71 |
ENST00000451655.5
|
BACH1
|
BTB domain and CNC homolog 1 |
| chr17_-_7252054 | 0.71 |
ENST00000575783.5
ENST00000573600.5 |
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr2_-_160200251 | 0.71 |
ENST00000428609.6
ENST00000409967.6 ENST00000283249.7 |
ITGB6
|
integrin subunit beta 6 |
| chr7_+_142554828 | 0.70 |
ENST00000611787.1
|
TRBV11-3
|
T cell receptor beta variable 11-3 |
| chr4_-_122621011 | 0.69 |
ENST00000611104.2
ENST00000648588.1 |
IL21
|
interleukin 21 |
| chr21_-_14546351 | 0.68 |
ENST00000619120.4
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr4_-_48114523 | 0.68 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr14_+_51847145 | 0.67 |
ENST00000615906.4
|
GNG2
|
G protein subunit gamma 2 |
| chr16_+_28984795 | 0.67 |
ENST00000395461.7
|
LAT
|
linker for activation of T cells |
| chr17_-_40799939 | 0.67 |
ENST00000306658.8
|
KRT28
|
keratin 28 |
| chr11_-_26572102 | 0.65 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated |
| chr3_-_155293665 | 0.65 |
ENST00000489090.2
|
STRIT1
|
small transmembrane regulator of ion transport 1 |
| chr12_+_20810698 | 0.65 |
ENST00000540853.5
ENST00000381545.8 |
SLCO1B3
|
solute carrier organic anion transporter family member 1B3 |
| chr9_-_35619542 | 0.65 |
ENST00000396757.6
|
CD72
|
CD72 molecule |
| chr10_-_6062290 | 0.65 |
ENST00000256876.10
ENST00000379954.5 |
IL2RA
|
interleukin 2 receptor subunit alpha |
| chr3_-_52454032 | 0.65 |
ENST00000232975.8
|
TNNC1
|
troponin C1, slow skeletal and cardiac type |
| chr5_-_95081858 | 0.64 |
ENST00000505465.1
|
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr1_-_222712428 | 0.64 |
ENST00000355727.3
ENST00000340020.11 |
AIDA
|
axin interactor, dorsalization associated |
| chr2_+_37342816 | 0.63 |
ENST00000650442.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr1_+_206440061 | 0.63 |
ENST00000604925.5
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr11_-_6619353 | 0.63 |
ENST00000642892.1
ENST00000645620.1 ENST00000533371.6 ENST00000647152.1 ENST00000644810.1 ENST00000299427.12 ENST00000682424.1 ENST00000644218.1 ENST00000528657.2 ENST00000531754.2 |
TPP1
|
tripeptidyl peptidase 1 |
| chr1_+_117001744 | 0.63 |
ENST00000256652.8
ENST00000682167.1 ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr6_+_63211446 | 0.63 |
ENST00000370659.1
|
FKBP1C
|
FKBP prolyl isomerase family member 1C |
| chr3_-_58210961 | 0.62 |
ENST00000486455.5
ENST00000394549.7 |
DNASE1L3
|
deoxyribonuclease 1 like 3 |
| chr20_+_45363454 | 0.62 |
ENST00000453003.1
|
SYS1
|
SYS1 golgi trafficking protein |
| chr5_-_180815528 | 0.62 |
ENST00000333055.8
|
MGAT1
|
alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase |
| chr19_-_54063882 | 0.61 |
ENST00000338372.7
|
VSTM1
|
V-set and transmembrane domain containing 1 |
| chr1_-_209802149 | 0.61 |
ENST00000456314.1
|
IRF6
|
interferon regulatory factor 6 |
| chr1_-_56966006 | 0.61 |
ENST00000371237.9
|
C8B
|
complement C8 beta chain |
| chr12_+_51239278 | 0.61 |
ENST00000551313.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr12_-_52814106 | 0.60 |
ENST00000551956.2
|
KRT4
|
keratin 4 |
| chr4_-_40630826 | 0.60 |
ENST00000505414.5
ENST00000511598.5 |
RBM47
|
RNA binding motif protein 47 |
| chr11_-_126268913 | 0.60 |
ENST00000532259.1
|
SRPRA
|
SRP receptor subunit alpha |
| chr3_-_146528750 | 0.59 |
ENST00000483300.5
|
PLSCR1
|
phospholipid scramblase 1 |
| chr2_-_160200310 | 0.59 |
ENST00000620391.4
|
ITGB6
|
integrin subunit beta 6 |
| chr16_+_3018390 | 0.59 |
ENST00000573001.5
|
TNFRSF12A
|
TNF receptor superfamily member 12A |
| chr3_-_12664101 | 0.59 |
ENST00000251849.9
ENST00000442415.7 |
RAF1
|
Raf-1 proto-oncogene, serine/threonine kinase |
| chr3_+_16265160 | 0.58 |
ENST00000627468.2
ENST00000605932.5 ENST00000435829.6 ENST00000285083.10 |
OXNAD1
|
oxidoreductase NAD binding domain containing 1 |
| chr3_-_27484335 | 0.58 |
ENST00000454389.5
ENST00000440156.5 ENST00000437179.5 ENST00000446700.5 ENST00000455077.5 |
SLC4A7
|
solute carrier family 4 member 7 |
| chr8_-_123653794 | 0.58 |
ENST00000684634.1
|
KLHL38
|
kelch like family member 38 |
| chr3_-_27484374 | 0.58 |
ENST00000445684.5
ENST00000425128.6 |
SLC4A7
|
solute carrier family 4 member 7 |
| chr1_-_99766620 | 0.57 |
ENST00000646001.2
|
FRRS1
|
ferric chelate reductase 1 |
| chr1_-_157841800 | 0.57 |
ENST00000368174.5
|
CD5L
|
CD5 molecule like |
| chr9_-_114388020 | 0.57 |
ENST00000312033.3
|
AKNA
|
AT-hook transcription factor |
| chr8_+_127735511 | 0.57 |
ENST00000517291.2
|
MYC
|
MYC proto-oncogene, bHLH transcription factor |
| chr12_-_102481744 | 0.56 |
ENST00000644491.1
|
IGF1
|
insulin like growth factor 1 |
| chr4_-_40630739 | 0.56 |
ENST00000511902.5
ENST00000505220.1 |
RBM47
|
RNA binding motif protein 47 |
| chr10_-_72088504 | 0.56 |
ENST00000536168.2
|
SPOCK2
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 2 |
| chr2_+_161231078 | 0.56 |
ENST00000439442.1
|
TANK
|
TRAF family member associated NFKB activator |
| chr14_+_24310134 | 0.56 |
ENST00000533293.2
ENST00000543919.1 |
LTB4R2
|
leukotriene B4 receptor 2 |
| chr19_+_43596480 | 0.56 |
ENST00000533118.5
|
ZNF576
|
zinc finger protein 576 |
| chr11_-_26572130 | 0.56 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr17_+_7252237 | 0.55 |
ENST00000570500.5
|
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr8_+_75539893 | 0.55 |
ENST00000674002.1
|
HNF4G
|
hepatocyte nuclear factor 4 gamma |
| chr5_+_134371561 | 0.55 |
ENST00000265339.7
ENST00000506787.5 ENST00000507277.1 |
UBE2B
|
ubiquitin conjugating enzyme E2 B |
| chr4_+_70518563 | 0.54 |
ENST00000339336.9
|
AMTN
|
amelotin |
| chr15_+_80072559 | 0.54 |
ENST00000560228.5
ENST00000559835.5 ENST00000559775.5 ENST00000558688.5 ENST00000560392.5 ENST00000560976.5 ENST00000558272.5 ENST00000558390.5 |
ZFAND6
|
zinc finger AN1-type containing 6 |
| chr13_+_48256214 | 0.54 |
ENST00000650237.1
|
ITM2B
|
integral membrane protein 2B |
| chr1_-_154206327 | 0.54 |
ENST00000368525.4
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr5_-_150086511 | 0.54 |
ENST00000675795.1
|
CSF1R
|
colony stimulating factor 1 receptor |
| chrX_-_30577759 | 0.54 |
ENST00000378962.4
|
TASL
|
TLR adaptor interacting with endolysosomal SLC15A4 |
| chr7_+_142645938 | 0.54 |
ENST00000390396.1
|
TRBV23-1
|
T cell receptor beta variable 23-1 (non-functional) |
| chr7_+_142399860 | 0.53 |
ENST00000390364.3
|
TRBV10-1
|
T cell receptor beta variable 10-1 |
| chr10_+_35195843 | 0.53 |
ENST00000488741.5
ENST00000474931.5 ENST00000468236.5 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFE2L1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.7 | 4.0 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.6 | 3.0 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.6 | 7.8 | GO:2000332 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.6 | 2.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.6 | 1.7 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.5 | 7.5 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.5 | 2.0 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.4 | 1.3 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.4 | 1.5 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.4 | 0.4 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.4 | 1.9 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.4 | 1.1 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.3 | 1.7 | GO:1902568 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.3 | 0.8 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.3 | 1.9 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.3 | 0.8 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.3 | 0.8 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.3 | 1.0 | GO:0044145 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.3 | 2.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.3 | 0.8 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.2 | 1.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 0.7 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.2 | 2.4 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.2 | 2.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 1.3 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.2 | 1.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 0.6 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.2 | 1.4 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.2 | 1.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.2 | 1.4 | GO:0090095 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.2 | 1.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 | 1.8 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.2 | 2.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.2 | 0.2 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) |
| 0.2 | 1.5 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 1.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.2 | 2.4 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 0.6 | GO:0002086 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 1.8 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.2 | 0.9 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.2 | 4.3 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.2 | 1.4 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 0.7 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.4 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.5 | GO:0060903 | regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of meiosis I(GO:0060903) |
| 0.1 | 0.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.4 | GO:0090108 | positive regulation of high-density lipoprotein particle assembly(GO:0090108) positive regulation of secretion of lysosomal enzymes(GO:0090340) |
| 0.1 | 1.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 1.3 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.6 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.5 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.1 | 0.4 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) |
| 0.1 | 0.5 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 1.2 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.5 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.3 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.7 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.9 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.4 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.1 | 1.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.6 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) |
| 0.1 | 0.7 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 0.9 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 1.4 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.4 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.1 | 0.3 | GO:0060279 | negative regulation of B cell differentiation(GO:0045578) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 1.4 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.3 | GO:1904882 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.5 | GO:1903758 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 0.4 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.5 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 | 0.3 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.6 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.4 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 1.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 0.2 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.1 | 1.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 3.0 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.7 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.2 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 1.5 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 2.3 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.8 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.2 | GO:0060003 | copper ion export(GO:0060003) |
| 0.0 | 0.6 | GO:0048296 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.0 | GO:0072343 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.0 | 0.8 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 1.4 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 1.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.5 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.9 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.5 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.5 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.4 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.3 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.5 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.3 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 1.5 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.6 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.5 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.3 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.3 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.7 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 1.0 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.8 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.4 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.4 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.8 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 1.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.5 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.4 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.7 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.0 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 1.2 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.6 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.4 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.1 | GO:0044349 | nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) DNA 3' dephosphorylation(GO:0098503) DNA 3' dephosphorylation involved in DNA repair(GO:0098504) polynucleotide 3' dephosphorylation(GO:0098506) |
| 0.0 | 2.3 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.0 | 0.1 | GO:1901804 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.9 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.2 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.5 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.3 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 1.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 2.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.4 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.2 | GO:0071964 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 2.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 3.4 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 3.6 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.1 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0051659 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 1.1 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.3 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.4 | GO:0071402 | cellular response to lipoprotein particle stimulus(GO:0071402) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 1.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:2000870 | oocyte growth(GO:0001555) regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.5 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.3 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.9 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.5 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0031622 | regulation of fever generation(GO:0031620) positive regulation of fever generation(GO:0031622) |
| 0.0 | 2.3 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.9 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.4 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.5 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 2.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.7 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 0.1 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.3 | GO:0060046 | fusion of sperm to egg plasma membrane(GO:0007342) regulation of acrosome reaction(GO:0060046) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 2.2 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.4 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 1.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0042423 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.0 | 0.9 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.5 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.4 | 1.8 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.4 | 1.3 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.4 | 1.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 1.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.3 | 1.3 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.2 | 1.7 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.2 | 2.3 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 1.0 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.2 | 0.5 | GO:0002141 | stereocilia ankle link(GO:0002141) USH2 complex(GO:1990696) |
| 0.2 | 7.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.7 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.4 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 1.7 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 0.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.5 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 1.9 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 2.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.3 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 1.0 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.4 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.7 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.3 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 1.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 2.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.5 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 5.2 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 4.3 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.8 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.7 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 3.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 2.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 1.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 7.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.4 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 3.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 1.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.6 | 7.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.4 | 2.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.4 | 4.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.4 | 1.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.3 | 1.7 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.3 | 1.9 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.3 | 2.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.3 | 0.8 | GO:0004411 | homogentisate 1,2-dioxygenase activity(GO:0004411) |
| 0.2 | 3.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 1.7 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 1.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.2 | 0.6 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.2 | 1.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 1.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 1.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 0.7 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.2 | 8.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.4 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.2 | 2.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 2.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.5 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.1 | 0.5 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.4 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.1 | 0.8 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.4 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.5 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 0.5 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 1.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 1.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 1.8 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.5 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 0.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.4 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.4 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 2.0 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.6 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 1.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.7 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.2 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 1.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.8 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 3.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 2.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 1.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 1.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 2.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.6 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 3.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.5 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.2 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.1 | 1.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 1.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.8 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.3 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 2.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.5 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 1.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.4 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.8 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.8 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 1.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 4.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 1.0 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.1 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.7 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 5.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 1.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.9 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.6 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 2.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 2.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 1.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 2.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.2 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 1.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 2.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.0 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 0.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 1.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 3.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 3.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 3.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 3.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 3.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 3.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 4.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 3.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 3.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 2.4 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 1.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 10.3 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 2.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.9 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 0.3 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 1.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 3.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.7 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 2.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 2.0 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 4.1 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.4 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 1.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 2.4 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 2.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 5.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.9 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |