Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
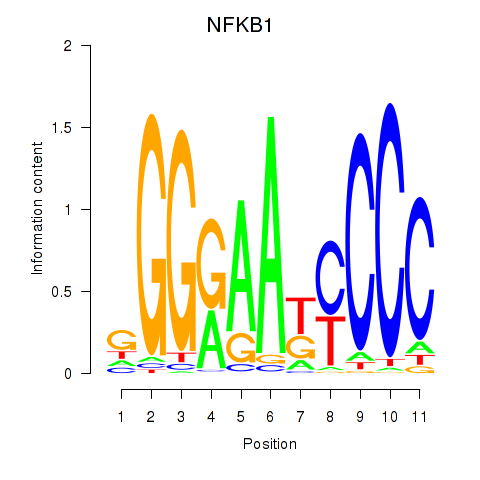
Results for NFKB1
Z-value: 1.53
Transcription factors associated with NFKB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFKB1
|
ENSG00000109320.13 | nuclear factor kappa B subunit 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFKB1 | hg38_v1_chr4_+_102501298_102501474 | 0.10 | 5.9e-01 | Click! |
Activity profile of NFKB1 motif
Sorted Z-values of NFKB1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFKB1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 1.9 | 7.5 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 1.3 | 4.0 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 1.0 | 3.1 | GO:0070446 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 1.0 | 4.0 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 1.0 | 2.9 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.8 | 3.1 | GO:0032499 | detection of peptidoglycan(GO:0032499) activation of MAPK activity involved in innate immune response(GO:0035419) |
| 0.8 | 8.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.7 | 2.8 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.7 | 2.0 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.6 | 2.5 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.6 | 1.8 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.6 | 1.2 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.6 | 1.7 | GO:0071206 | establishment of protein localization to juxtaparanode region of axon(GO:0071206) |
| 0.6 | 1.7 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.5 | 1.6 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.5 | 1.6 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.5 | 6.0 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.5 | 1.6 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.5 | 3.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.5 | 3.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.5 | 11.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.5 | 2.4 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.5 | 1.4 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.5 | 4.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.4 | 2.2 | GO:0038156 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.4 | 2.6 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.4 | 3.6 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.4 | 3.6 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.4 | 1.1 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.4 | 7.5 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.4 | 2.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.3 | 1.0 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.3 | 1.0 | GO:0002730 | regulation of dendritic cell cytokine production(GO:0002730) |
| 0.3 | 2.0 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.3 | 3.6 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.3 | 2.0 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.3 | 2.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.3 | 3.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.3 | 1.8 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.3 | 0.9 | GO:0060516 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.3 | 1.5 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.3 | 1.2 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.3 | 2.3 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.3 | 0.8 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.3 | 2.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.3 | 1.3 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.3 | 2.4 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.3 | 1.3 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.2 | 2.7 | GO:1904327 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.2 | 1.4 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.2 | 4.7 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.2 | 2.4 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 1.7 | GO:0045357 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.2 | 0.6 | GO:1904907 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.2 | 2.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 1.7 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.2 | 2.7 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.2 | 1.0 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.2 | 1.8 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.2 | 0.6 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.2 | 0.6 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.2 | 1.0 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.2 | 1.5 | GO:0023035 | CD40 signaling pathway(GO:0023035) protein linear polyubiquitination(GO:0097039) |
| 0.2 | 1.7 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.2 | 0.9 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 1.6 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.2 | 3.6 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.2 | 0.6 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 1.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 1.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 2.3 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 1.0 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 0.4 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 0.1 | 1.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.6 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 2.9 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 1.3 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 1.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 2.8 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 2.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 11.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.8 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.1 | 0.7 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.1 | 1.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 1.8 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 1.6 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 1.8 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.2 | GO:0035397 | helper T cell enhancement of adaptive immune response(GO:0035397) |
| 0.1 | 0.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.3 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 0.6 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.1 | 0.4 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 1.8 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 0.3 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 0.3 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.1 | 1.0 | GO:0010819 | regulation of T cell chemotaxis(GO:0010819) |
| 0.1 | 0.9 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.4 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 2.0 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 1.0 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.1 | 0.3 | GO:1903755 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.1 | 0.2 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 2.8 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 1.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.4 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 4.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 0.7 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 2.1 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.7 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.5 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.9 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.5 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 1.5 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 2.0 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.3 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.1 | 0.4 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 1.4 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 1.5 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 1.6 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 7.9 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 1.6 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.1 | 5.0 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 1.0 | GO:0033327 | luteinization(GO:0001553) Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 1.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 2.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.5 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.8 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 0.1 | GO:1990619 | histone H3-K9 deacetylation(GO:1990619) |
| 0.0 | 0.7 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 1.6 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.3 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 1.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.5 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.5 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.4 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.0 | 0.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.3 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 1.0 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 1.3 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.8 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 1.6 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 1.0 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.2 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 1.9 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.4 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 3.8 | GO:0090003 | regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.0 | 1.8 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.9 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 3.0 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 7.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 2.1 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.3 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 1.3 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 2.1 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.4 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 4.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 2.1 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.8 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 3.4 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.2 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.6 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 2.1 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 1.2 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.3 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 1.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 2.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 3.0 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 1.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.3 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 1.2 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.5 | GO:0002438 | acute inflammatory response to antigenic stimulus(GO:0002438) |
| 0.0 | 0.1 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.4 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 1.8 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.9 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.5 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.8 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.0 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.4 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.0 | 0.4 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.8 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.6 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.9 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 1.0 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.6 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 3.2 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.5 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.4 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.5 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 2.0 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 0.7 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 1.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 1.6 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.0 | 0.5 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.6 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.2 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 1.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0030526 | granulocyte macrophage colony-stimulating factor receptor complex(GO:0030526) |
| 0.6 | 2.5 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.5 | 2.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.5 | 5.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.4 | 3.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.4 | 2.9 | GO:0042825 | TAP complex(GO:0042825) |
| 0.4 | 8.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.3 | 3.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.3 | 2.6 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.3 | 1.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.3 | 5.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 1.4 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.3 | 2.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.2 | 2.7 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 1.5 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 0.7 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 1.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.2 | 2.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 0.6 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.2 | 2.1 | GO:0042611 | MHC protein complex(GO:0042611) MHC class I protein complex(GO:0042612) |
| 0.2 | 3.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 1.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 1.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 3.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 2.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 2.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.8 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 3.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 7.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 2.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.9 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 3.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 1.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 6.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 4.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 1.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 0.3 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.4 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.1 | 1.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.3 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.1 | 10.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.0 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.9 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 3.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.4 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.1 | 0.3 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 0.9 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.3 | GO:0070993 | translation preinitiation complex(GO:0070993) |
| 0.1 | 6.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 17.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 0.4 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 0.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.6 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 1.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 6.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 2.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 3.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.1 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 1.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 2.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.1 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.5 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 1.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 5.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 1.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 2.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 2.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.9 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 6.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.9 | GO:0034702 | ion channel complex(GO:0034702) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.9 | 8.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.8 | 2.3 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.6 | 4.6 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.6 | 1.7 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.5 | 2.0 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.4 | 2.2 | GO:0004914 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 0.4 | 1.3 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.4 | 8.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.4 | 2.8 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.3 | 3.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.3 | 1.6 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.3 | 2.6 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.3 | 1.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.3 | 1.5 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.3 | 16.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 0.8 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.3 | 2.4 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.3 | 4.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 2.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.3 | 7.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 2.0 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.2 | 4.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 1.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 0.8 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.2 | 1.0 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.2 | 1.8 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 1.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 10.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 0.3 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 1.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 3.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 1.6 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 1.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 2.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 4.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.2 | 2.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 2.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 2.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.4 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.1 | 0.7 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 0.7 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 2.0 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.1 | 0.4 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.1 | 2.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 2.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 12.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 2.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.6 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.1 | 0.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 1.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.8 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 1.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.8 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 2.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 1.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.4 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 0.9 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 3.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.8 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 0.4 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.1 | 1.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.6 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.3 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 1.5 | GO:0010857 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.1 | 0.2 | GO:0035877 | death effector domain binding(GO:0035877) cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 1.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.4 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.3 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.1 | 3.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 1.3 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.4 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 1.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 4.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 2.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 1.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 5.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 1.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 7.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.3 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 14.2 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 2.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 1.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 2.1 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 1.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 1.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 15.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 2.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 3.4 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 1.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.5 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0061598 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 1.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 2.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 6.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.2 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 1.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 2.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 4.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 5.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 11.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 9.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.2 | 8.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 5.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 8.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 1.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 2.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 1.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 3.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 2.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 4.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 8.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 3.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 2.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 1.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.8 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 2.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 3.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 5.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.8 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 2.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 2.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 2.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 3.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 3.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 3.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 2.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 18.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.3 | 8.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 7.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 1.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.2 | 8.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 4.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 5.7 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.2 | 2.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 7.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.7 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 3.0 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 4.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 2.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 2.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 4.5 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.1 | 2.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 1.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 2.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 5.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 1.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 2.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 7.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 3.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 5.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 5.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 3.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 2.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 4.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 2.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.7 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 1.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 2.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 2.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.2 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.0 | 2.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 3.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.2 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.9 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 3.4 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |