Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for NKX1-2_RAX
Z-value: 0.74
Transcription factors associated with NKX1-2_RAX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
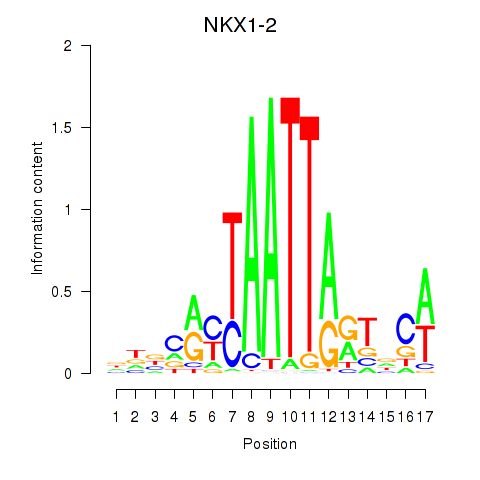
NKX1-2
|
ENSG00000229544.9 | NK1 homeobox 2 |
|
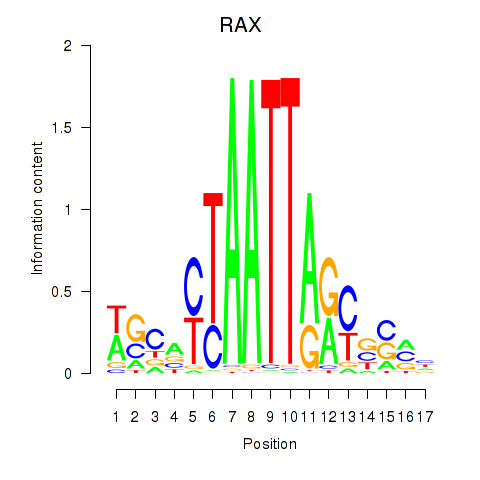
RAX
|
ENSG00000134438.10 | retina and anterior neural fold homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RAX | hg38_v1_chr18_-_59273379_59273429 | -0.45 | 1.0e-02 | Click! |
| NKX1-2 | hg38_v1_chr10_-_124450027_124450043 | -0.06 | 7.6e-01 | Click! |
Activity profile of NKX1-2_RAX motif
Sorted Z-values of NKX1-2_RAX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_115593570 | 2.70 |
ENST00000539310.5
|
PLS3
|
plastin 3 |
| chr12_-_21910853 | 2.06 |
ENST00000544039.5
|
ABCC9
|
ATP binding cassette subfamily C member 9 |
| chr12_-_91180365 | 2.01 |
ENST00000547937.5
|
DCN
|
decorin |
| chr1_+_186296267 | 1.72 |
ENST00000533951.5
ENST00000367482.8 ENST00000635041.1 ENST00000367483.8 ENST00000367485.4 ENST00000445192.7 |
PRG4
|
proteoglycan 4 |
| chrX_+_43656289 | 1.71 |
ENST00000338702.4
|
MAOA
|
monoamine oxidase A |
| chr10_+_97446194 | 1.68 |
ENST00000370846.8
ENST00000352634.8 ENST00000353979.7 ENST00000370842.6 ENST00000345745.9 |
ZDHHC16
|
zinc finger DHHC-type palmitoyltransferase 16 |
| chrX_-_21658324 | 1.65 |
ENST00000379499.3
|
KLHL34
|
kelch like family member 34 |
| chr6_+_125153649 | 1.54 |
ENST00000304877.17
ENST00000368402.9 ENST00000368388.6 ENST00000534000.6 |
TPD52L1
|
TPD52 like 1 |
| chr14_+_32329341 | 1.54 |
ENST00000557354.5
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A-kinase anchoring protein 6 |
| chr14_+_32329256 | 1.53 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6 |
| chr2_+_216659600 | 1.40 |
ENST00000456764.1
|
IGFBP2
|
insulin like growth factor binding protein 2 |
| chr13_-_110242694 | 1.37 |
ENST00000648989.1
ENST00000647797.1 ENST00000648966.1 ENST00000649484.1 ENST00000648695.1 ENST00000650115.1 ENST00000650566.1 |
COL4A1
|
collagen type IV alpha 1 chain |
| chr13_-_35476682 | 1.36 |
ENST00000379919.6
|
MAB21L1
|
mab-21 like 1 |
| chr6_+_54099565 | 1.29 |
ENST00000511678.5
|
MLIP
|
muscular LMNA interacting protein |
| chr9_-_92404559 | 1.27 |
ENST00000262551.8
ENST00000375561.10 |
OGN
|
osteoglycin |
| chr3_-_151316795 | 1.27 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr4_+_76435216 | 1.26 |
ENST00000296043.7
|
SHROOM3
|
shroom family member 3 |
| chr10_+_97446137 | 1.22 |
ENST00000370854.7
ENST00000393760.6 ENST00000414567.5 |
ZDHHC16
|
zinc finger DHHC-type palmitoyltransferase 16 |
| chr1_-_151831768 | 1.21 |
ENST00000318247.7
|
RORC
|
RAR related orphan receptor C |
| chr3_-_105869035 | 1.21 |
ENST00000447441.6
ENST00000403724.5 ENST00000405772.5 |
CBLB
|
Cbl proto-oncogene B |
| chr6_+_54099538 | 1.19 |
ENST00000447836.6
|
MLIP
|
muscular LMNA interacting protein |
| chr3_-_105868964 | 1.17 |
ENST00000394030.8
|
CBLB
|
Cbl proto-oncogene B |
| chr6_+_54018910 | 1.17 |
ENST00000514921.5
ENST00000274897.9 ENST00000370877.6 |
MLIP
|
muscular LMNA interacting protein |
| chr20_+_45416551 | 1.05 |
ENST00000639292.1
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis class T |
| chr17_+_1762814 | 1.04 |
ENST00000570731.5
|
SERPINF1
|
serpin family F member 1 |
| chr1_+_160400543 | 1.04 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr4_-_145180496 | 1.03 |
ENST00000447906.8
|
OTUD4
|
OTU deubiquitinase 4 |
| chr10_+_24466487 | 1.01 |
ENST00000396446.5
ENST00000396445.5 ENST00000376451.4 |
KIAA1217
|
KIAA1217 |
| chr16_-_55833085 | 0.99 |
ENST00000360526.8
|
CES1
|
carboxylesterase 1 |
| chr9_+_121567057 | 0.98 |
ENST00000394340.7
ENST00000436835.5 ENST00000259371.6 |
DAB2IP
|
DAB2 interacting protein |
| chr7_+_71132123 | 0.98 |
ENST00000333538.10
|
GALNT17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr2_-_189582012 | 0.95 |
ENST00000427419.5
ENST00000455320.5 |
SLC40A1
|
solute carrier family 40 member 1 |
| chr1_-_151831837 | 0.91 |
ENST00000652040.1
|
RORC
|
RAR related orphan receptor C |
| chr7_+_120988683 | 0.89 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr19_+_49487510 | 0.88 |
ENST00000679106.1
ENST00000621674.4 ENST00000391857.9 ENST00000678510.1 ENST00000467825.2 |
RPL13A
|
ribosomal protein L13a |
| chr16_-_55833186 | 0.82 |
ENST00000361503.8
ENST00000422046.6 |
CES1
|
carboxylesterase 1 |
| chr14_+_19719015 | 0.78 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor family 4 subfamily N member 2 |
| chr6_-_166627244 | 0.77 |
ENST00000265678.9
|
RPS6KA2
|
ribosomal protein S6 kinase A2 |
| chr7_+_101127095 | 0.76 |
ENST00000223095.5
|
SERPINE1
|
serpin family E member 1 |
| chr3_+_186842687 | 0.75 |
ENST00000444204.2
ENST00000320741.7 |
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr3_-_120682113 | 0.74 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr11_-_18236795 | 0.74 |
ENST00000278222.7
|
SAA4
|
serum amyloid A4, constitutive |
| chr1_+_162366896 | 0.74 |
ENST00000420220.1
|
ENSG00000254706.3
|
novel protein |
| chr2_-_219399981 | 0.73 |
ENST00000519905.1
ENST00000523282.5 ENST00000434339.5 ENST00000457935.5 |
DNPEP
|
aspartyl aminopeptidase |
| chr3_-_105869387 | 0.70 |
ENST00000438603.6
ENST00000443752.2 |
CBLB
|
Cbl proto-oncogene B |
| chr5_+_67004618 | 0.66 |
ENST00000261569.11
ENST00000436277.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr7_-_7535863 | 0.65 |
ENST00000399429.8
|
COL28A1
|
collagen type XXVIII alpha 1 chain |
| chr6_+_132538290 | 0.64 |
ENST00000434551.2
|
TAAR9
|
trace amine associated receptor 9 |
| chr5_-_35938572 | 0.62 |
ENST00000651391.1
ENST00000397366.5 ENST00000513623.5 ENST00000514524.2 ENST00000397367.6 |
CAPSL
|
calcyphosine like |
| chr11_+_112176364 | 0.62 |
ENST00000526088.5
ENST00000532593.5 ENST00000531169.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr12_+_6904962 | 0.61 |
ENST00000415834.5
ENST00000436789.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr17_-_7263959 | 0.59 |
ENST00000571932.2
|
CLDN7
|
claudin 7 |
| chr12_+_6904733 | 0.59 |
ENST00000007969.12
ENST00000622489.4 ENST00000443597.7 ENST00000323702.9 |
LRRC23
|
leucine rich repeat containing 23 |
| chr16_+_82034978 | 0.59 |
ENST00000563491.5
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2 |
| chr1_-_16978276 | 0.58 |
ENST00000375534.7
|
MFAP2
|
microfibril associated protein 2 |
| chr19_-_6424772 | 0.57 |
ENST00000619396.4
ENST00000398148.7 |
KHSRP
|
KH-type splicing regulatory protein |
| chr3_+_184135343 | 0.56 |
ENST00000648915.2
ENST00000432569.2 ENST00000647909.1 |
EIF2B5
|
eukaryotic translation initiation factor 2B subunit epsilon |
| chr1_-_159862648 | 0.52 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr7_-_107803215 | 0.52 |
ENST00000340010.10
ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 member 3 |
| chr1_+_42682388 | 0.52 |
ENST00000321358.12
ENST00000332220.10 |
YBX1
|
Y-box binding protein 1 |
| chr5_-_103562775 | 0.51 |
ENST00000230792.7
ENST00000507423.1 |
NUDT12
|
nudix hydrolase 12 |
| chr19_-_3985451 | 0.51 |
ENST00000309311.7
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr19_+_48445795 | 0.51 |
ENST00000598711.1
|
GRWD1
|
glutamate rich WD repeat containing 1 |
| chr11_-_125111708 | 0.51 |
ENST00000531909.5
ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chrX_+_19343893 | 0.50 |
ENST00000540249.5
ENST00000545074.5 ENST00000379806.9 ENST00000423505.5 ENST00000422285.7 ENST00000417819.5 ENST00000355808.9 ENST00000379805.3 |
PDHA1
|
pyruvate dehydrogenase E1 subunit alpha 1 |
| chr11_-_125111579 | 0.50 |
ENST00000532156.5
ENST00000532407.5 ENST00000279968.8 ENST00000527766.5 ENST00000529583.5 ENST00000524373.5 ENST00000527271.5 ENST00000526175.5 ENST00000529609.5 ENST00000682305.1 ENST00000533273.1 |
TMEM218
|
transmembrane protein 218 |
| chr11_-_59615673 | 0.48 |
ENST00000263847.6
|
OSBP
|
oxysterol binding protein |
| chr6_+_30720335 | 0.47 |
ENST00000327892.13
|
TUBB
|
tubulin beta class I |
| chr11_+_72189528 | 0.47 |
ENST00000312293.9
|
FOLR1
|
folate receptor alpha |
| chr4_-_102827948 | 0.46 |
ENST00000394804.6
ENST00000394801.8 |
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr19_-_45782479 | 0.46 |
ENST00000447742.6
ENST00000354227.9 ENST00000291270.9 ENST00000683086.1 ENST00000343373.10 |
DMPK
|
DM1 protein kinase |
| chr7_+_138460238 | 0.46 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24 |
| chr14_-_99480831 | 0.45 |
ENST00000331768.10
ENST00000630307.2 |
SETD3
|
SET domain containing 3, actin histidine methyltransferase |
| chr5_-_95554944 | 0.45 |
ENST00000513823.5
ENST00000649566.1 ENST00000358746.7 ENST00000514952.5 |
TTC37
|
tetratricopeptide repeat domain 37 |
| chr12_+_26195313 | 0.44 |
ENST00000422622.3
|
SSPN
|
sarcospan |
| chr6_-_111606260 | 0.43 |
ENST00000340026.10
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr8_-_42768602 | 0.43 |
ENST00000534622.5
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit |
| chr6_-_31862809 | 0.42 |
ENST00000375631.5
|
NEU1
|
neuraminidase 1 |
| chr4_-_102827723 | 0.42 |
ENST00000349311.12
|
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr4_-_102828159 | 0.41 |
ENST00000394803.9
|
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chrX_+_136536099 | 0.40 |
ENST00000440515.5
ENST00000456412.1 |
VGLL1
|
vestigial like family member 1 |
| chr5_-_83673544 | 0.39 |
ENST00000503117.1
ENST00000510978.5 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr3_+_28349146 | 0.39 |
ENST00000420223.5
|
ZCWPW2
|
zinc finger CW-type and PWWP domain containing 2 |
| chr13_-_109786567 | 0.37 |
ENST00000375856.5
|
IRS2
|
insulin receptor substrate 2 |
| chr19_-_9107475 | 0.37 |
ENST00000641081.1
|
OR7G2
|
olfactory receptor family 7 subfamily G member 2 |
| chr16_-_20691256 | 0.37 |
ENST00000307493.8
|
ACSM1
|
acyl-CoA synthetase medium chain family member 1 |
| chr11_+_72189659 | 0.37 |
ENST00000393681.6
|
FOLR1
|
folate receptor alpha |
| chr9_+_116687295 | 0.37 |
ENST00000450136.2
ENST00000373983.2 ENST00000411410.1 |
TRIM32
|
tripartite motif containing 32 |
| chr2_-_238288600 | 0.37 |
ENST00000254657.8
|
PER2
|
period circadian regulator 2 |
| chr10_-_97445850 | 0.36 |
ENST00000477692.6
ENST00000485122.6 ENST00000370886.9 ENST00000370885.8 ENST00000370902.8 ENST00000370884.5 |
EXOSC1
|
exosome component 1 |
| chr1_-_211134135 | 0.36 |
ENST00000638983.1
ENST00000271751.10 ENST00000639952.1 |
ENSG00000284299.1
KCNH1
|
novel protein potassium voltage-gated channel subfamily H member 1 |
| chr6_-_111605859 | 0.36 |
ENST00000651359.1
ENST00000650859.1 ENST00000359831.8 ENST00000368761.11 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr15_-_43589942 | 0.36 |
ENST00000429176.5
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr9_+_122370523 | 0.35 |
ENST00000643810.1
ENST00000540753.6 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chrM_+_9207 | 0.35 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr17_-_5035418 | 0.35 |
ENST00000254853.10
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 member 1 |
| chr1_-_211134061 | 0.34 |
ENST00000639602.1
ENST00000638498.1 ENST00000367007.5 |
ENSG00000284299.1
KCNH1
|
novel protein potassium voltage-gated channel subfamily H member 1 |
| chr6_+_160121859 | 0.33 |
ENST00000324965.8
ENST00000457470.6 |
SLC22A1
|
solute carrier family 22 member 1 |
| chr12_-_9869345 | 0.33 |
ENST00000228438.3
|
CLEC2B
|
C-type lectin domain family 2 member B |
| chr19_-_6424802 | 0.33 |
ENST00000600480.2
|
KHSRP
|
KH-type splicing regulatory protein |
| chr4_-_102828022 | 0.32 |
ENST00000502690.5
|
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr14_-_52791597 | 0.31 |
ENST00000216410.8
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr9_-_5339874 | 0.31 |
ENST00000223862.2
|
RLN1
|
relaxin 1 |
| chr11_-_4368386 | 0.31 |
ENST00000624801.3
|
OR52B4
|
olfactory receptor family 52 subfamily B member 4 |
| chr17_-_10469558 | 0.31 |
ENST00000255381.2
|
MYH4
|
myosin heavy chain 4 |
| chr20_+_45420585 | 0.31 |
ENST00000639239.1
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis class T |
| chr12_+_64405046 | 0.30 |
ENST00000540203.5
|
XPOT
|
exportin for tRNA |
| chr15_+_101812202 | 0.30 |
ENST00000332238.5
|
OR4F15
|
olfactory receptor family 4 subfamily F member 15 |
| chr14_-_99480784 | 0.30 |
ENST00000329331.7
ENST00000436070.6 |
SETD3
|
SET domain containing 3, actin histidine methyltransferase |
| chr11_-_122116215 | 0.29 |
ENST00000560104.2
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr18_+_58341038 | 0.29 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr10_-_104085793 | 0.29 |
ENST00000650263.1
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr19_-_45782388 | 0.28 |
ENST00000458663.6
|
DMPK
|
DM1 protein kinase |
| chr12_+_16347637 | 0.27 |
ENST00000543076.5
ENST00000396210.8 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr11_+_118527463 | 0.27 |
ENST00000302783.10
|
TTC36
|
tetratricopeptide repeat domain 36 |
| chr9_+_72577788 | 0.27 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1 |
| chr10_-_104085847 | 0.27 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr21_-_31038476 | 0.27 |
ENST00000382822.2
|
KRTAP19-8
|
keratin associated protein 19-8 |
| chr4_-_102828048 | 0.27 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr17_-_42185452 | 0.26 |
ENST00000293330.1
|
HCRT
|
hypocretin neuropeptide precursor |
| chr11_+_19712823 | 0.25 |
ENST00000396085.6
ENST00000349880.9 |
NAV2
|
neuron navigator 2 |
| chr10_-_104085762 | 0.25 |
ENST00000393211.3
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr6_-_82247697 | 0.25 |
ENST00000306270.12
ENST00000610980.4 |
IBTK
|
inhibitor of Bruton tyrosine kinase |
| chr12_+_8157034 | 0.24 |
ENST00000396570.7
|
ZNF705A
|
zinc finger protein 705A |
| chr11_-_111871530 | 0.24 |
ENST00000614444.4
ENST00000616540.5 |
ALG9
|
ALG9 alpha-1,2-mannosyltransferase |
| chr19_+_35748549 | 0.23 |
ENST00000301159.14
|
LIN37
|
lin-37 DREAM MuvB core complex component |
| chr9_+_34646654 | 0.23 |
ENST00000378842.8
|
GALT
|
galactose-1-phosphate uridylyltransferase |
| chr17_+_7407838 | 0.23 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2 |
| chr14_+_67364849 | 0.23 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2 subunit alpha |
| chr13_+_35476740 | 0.23 |
ENST00000537702.5
|
NBEA
|
neurobeachin |
| chr5_-_126595237 | 0.23 |
ENST00000637206.1
ENST00000553117.5 |
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1 |
| chr4_-_73620500 | 0.22 |
ENST00000335049.5
|
RASSF6
|
Ras association domain family member 6 |
| chr11_-_56748696 | 0.22 |
ENST00000641581.1
ENST00000641668.1 |
OR9G4
|
olfactory receptor family 9 subfamily G member 4 |
| chr6_+_154039333 | 0.22 |
ENST00000428397.6
|
OPRM1
|
opioid receptor mu 1 |
| chr8_-_96261579 | 0.22 |
ENST00000517720.1
ENST00000523821.5 ENST00000287025.4 |
MTERF3
|
mitochondrial transcription termination factor 3 |
| chr9_-_21240002 | 0.22 |
ENST00000380222.4
|
IFNA14
|
interferon alpha 14 |
| chr19_+_49766962 | 0.22 |
ENST00000354293.10
ENST00000359032.9 |
AP2A1
|
adaptor related protein complex 2 subunit alpha 1 |
| chr4_-_73620629 | 0.21 |
ENST00000342081.7
|
RASSF6
|
Ras association domain family member 6 |
| chr2_+_90038848 | 0.21 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr11_+_124183219 | 0.21 |
ENST00000641351.2
|
OR10D3
|
olfactory receptor family 10 subfamily D member 3 |
| chr9_+_122371036 | 0.20 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr7_+_100049765 | 0.20 |
ENST00000456748.6
ENST00000292450.9 ENST00000438937.1 ENST00000543588.2 |
ZSCAN21
|
zinc finger and SCAN domain containing 21 |
| chr3_+_195720867 | 0.20 |
ENST00000436408.6
|
MUC20
|
mucin 20, cell surface associated |
| chr7_-_100119291 | 0.20 |
ENST00000431404.2
|
TAF6
|
TATA-box binding protein associated factor 6 |
| chr9_+_34646589 | 0.20 |
ENST00000450095.6
|
GALT
|
galactose-1-phosphate uridylyltransferase |
| chrX_+_77899440 | 0.20 |
ENST00000373335.4
ENST00000475465.1 ENST00000650309.2 ENST00000647835.1 |
COX7B
|
cytochrome c oxidase subunit 7B |
| chr1_+_27934980 | 0.20 |
ENST00000373894.8
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr20_-_1466904 | 0.20 |
ENST00000476071.5
|
NSFL1C
|
NSFL1 cofactor |
| chr7_-_100119840 | 0.20 |
ENST00000437822.6
|
TAF6
|
TATA-box binding protein associated factor 6 |
| chr7_+_21543020 | 0.19 |
ENST00000409508.8
ENST00000620169.4 ENST00000328843.10 |
DNAH11
|
dynein axonemal heavy chain 11 |
| chr20_-_1466822 | 0.19 |
ENST00000353088.6
ENST00000216879.9 |
NSFL1C
|
NSFL1 cofactor |
| chr11_-_111871271 | 0.19 |
ENST00000398006.6
|
ALG9
|
ALG9 alpha-1,2-mannosyltransferase |
| chr4_-_106368772 | 0.18 |
ENST00000638719.4
|
GIMD1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr17_-_4967790 | 0.18 |
ENST00000575142.5
ENST00000206020.8 |
SPAG7
|
sperm associated antigen 7 |
| chr12_+_18261511 | 0.18 |
ENST00000538779.6
ENST00000433979.6 |
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr7_-_100119323 | 0.18 |
ENST00000523306.5
ENST00000344095.8 ENST00000417349.5 ENST00000493322.5 ENST00000520135.5 ENST00000460673.2 ENST00000453269.7 ENST00000452041.5 ENST00000452438.6 ENST00000451699.5 |
TAF6
|
TATA-box binding protein associated factor 6 |
| chr5_+_132873660 | 0.17 |
ENST00000296877.3
|
LEAP2
|
liver enriched antimicrobial peptide 2 |
| chr12_-_10807286 | 0.17 |
ENST00000240615.3
|
TAS2R8
|
taste 2 receptor member 8 |
| chr8_-_100559702 | 0.17 |
ENST00000520311.5
ENST00000520552.5 ENST00000521345.1 ENST00000523000.5 ENST00000335659.7 ENST00000358990.3 ENST00000519597.5 |
ANKRD46
|
ankyrin repeat domain 46 |
| chr7_-_100119317 | 0.17 |
ENST00000440225.5
|
TAF6
|
TATA-box binding protein associated factor 6 |
| chr9_+_34646627 | 0.16 |
ENST00000556278.1
|
ENSG00000258728.1
|
novel protein (GALT-IL11RA readthrough) |
| chr9_+_34329545 | 0.15 |
ENST00000379158.7
|
NUDT2
|
nudix hydrolase 2 |
| chr21_-_7793954 | 0.15 |
ENST00000624951.1
|
SMIM34B
|
small integral membrane protein 34B |
| chr21_-_36542600 | 0.15 |
ENST00000399136.5
|
CLDN14
|
claudin 14 |
| chr1_+_54715837 | 0.15 |
ENST00000371281.4
|
TTC4
|
tetratricopeptide repeat domain 4 |
| chr19_+_37317929 | 0.15 |
ENST00000589801.1
|
ZNF875
|
zinc finger protein 875 |
| chr9_+_72577369 | 0.15 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr4_-_67963441 | 0.13 |
ENST00000508048.6
|
TMPRSS11A
|
transmembrane serine protease 11A |
| chr19_+_8630622 | 0.13 |
ENST00000671902.1
ENST00000673603.1 |
NFILZ
|
NFIL3 like basic leucine zipper |
| chr9_-_21187671 | 0.12 |
ENST00000421715.2
|
IFNA4
|
interferon alpha 4 |
| chrX_-_41665766 | 0.12 |
ENST00000643043.2
ENST00000486402.1 ENST00000646087.2 |
CASK
|
calcium/calmodulin dependent serine protein kinase |
| chr15_+_49170200 | 0.12 |
ENST00000396509.6
|
GALK2
|
galactokinase 2 |
| chr11_+_28108248 | 0.12 |
ENST00000406787.7
ENST00000403099.5 ENST00000407364.8 |
METTL15
|
methyltransferase like 15 |
| chr8_-_81461574 | 0.12 |
ENST00000379071.4
|
FABP9
|
fatty acid binding protein 9 |
| chr12_-_55296569 | 0.11 |
ENST00000358433.3
|
OR6C6
|
olfactory receptor family 6 subfamily C member 6 |
| chr15_+_49170237 | 0.11 |
ENST00000560031.6
ENST00000558145.5 ENST00000544523.5 ENST00000560138.5 |
GALK2
|
galactokinase 2 |
| chr9_+_122371014 | 0.11 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr3_-_11643871 | 0.11 |
ENST00000430365.7
|
VGLL4
|
vestigial like family member 4 |
| chr1_-_152414256 | 0.11 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chrX_+_152831054 | 0.10 |
ENST00000370274.8
|
NSDHL
|
NAD(P) dependent steroid dehydrogenase-like |
| chr18_+_24460630 | 0.10 |
ENST00000256906.5
|
HRH4
|
histamine receptor H4 |
| chr8_+_9555900 | 0.10 |
ENST00000310430.11
ENST00000520408.5 ENST00000522110.1 |
TNKS
|
tankyrase |
| chr2_+_74455077 | 0.10 |
ENST00000431187.5
ENST00000233331.12 ENST00000409917.5 ENST00000409493.3 |
INO80B
|
INO80 complex subunit B |
| chr1_+_45688165 | 0.10 |
ENST00000372025.5
|
TMEM69
|
transmembrane protein 69 |
| chr11_+_92969651 | 0.09 |
ENST00000257068.3
ENST00000528076.1 |
MTNR1B
|
melatonin receptor 1B |
| chr14_+_55027200 | 0.09 |
ENST00000395472.2
ENST00000555846.2 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr6_-_93419545 | 0.09 |
ENST00000369297.1
ENST00000369303.9 ENST00000680224.1 ENST00000681532.1 ENST00000679565.1 |
EPHA7
|
EPH receptor A7 |
| chr22_-_28094135 | 0.08 |
ENST00000442232.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr1_+_192636121 | 0.08 |
ENST00000543215.5
ENST00000391995.7 |
RGS13
|
regulator of G protein signaling 13 |
| chr6_+_49499965 | 0.08 |
ENST00000545705.1
|
GLYATL3
|
glycine-N-acyltransferase like 3 |
| chr13_+_48037692 | 0.08 |
ENST00000258662.3
|
NUDT15
|
nudix hydrolase 15 |
| chr10_+_18260715 | 0.07 |
ENST00000615785.4
ENST00000617363.4 ENST00000396576.6 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr10_+_35605337 | 0.07 |
ENST00000321660.2
|
GJD4
|
gap junction protein delta 4 |
| chr17_-_73262352 | 0.07 |
ENST00000397671.1
|
CPSF4L
|
cleavage and polyadenylation specific factor 4 like |
| chr8_+_104223344 | 0.07 |
ENST00000523362.5
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chrX_-_152830721 | 0.06 |
ENST00000370277.5
|
CETN2
|
centrin 2 |
| chr18_+_24460655 | 0.06 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr1_-_158426237 | 0.06 |
ENST00000641042.1
|
OR10K2
|
olfactory receptor family 10 subfamily K member 2 |
| chr9_-_134068012 | 0.06 |
ENST00000303407.12
|
BRD3
|
bromodomain containing 3 |
| chr21_-_34423951 | 0.06 |
ENST00000450895.2
|
SMIM34A
|
small integral membrane protein 34A |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX1-2_RAX
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.3 | 3.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.3 | 1.0 | GO:1903988 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.3 | 0.8 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.3 | 1.4 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.3 | 1.0 | GO:0060448 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.3 | 0.8 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.2 | 0.7 | GO:0072244 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.2 | 0.6 | GO:0046247 | tetraterpenoid metabolic process(GO:0016108) carotenoid metabolic process(GO:0016116) carotene catabolic process(GO:0016121) xanthophyll metabolic process(GO:0016122) terpene catabolic process(GO:0046247) |
| 0.2 | 0.5 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.2 | 3.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.4 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 3.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.0 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 1.7 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.4 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 0.3 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 1.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 2.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.6 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 0.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 1.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 0.4 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 0.4 | GO:0051941 | lactate biosynthetic process(GO:0019249) regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 | 0.9 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 2.1 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.1 | 1.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 2.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.8 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.1 | 0.2 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.5 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.5 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.9 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 1.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.3 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.5 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.7 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 1.4 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.5 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.4 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:1904907 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.0 | 0.2 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.3 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.0 | 0.5 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 2.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.8 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 1.9 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 1.0 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.5 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 1.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 2.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 1.0 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.2 | 3.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 2.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 0.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.4 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.9 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.4 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 0.4 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.2 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) translation initiation ternary complex(GO:0044207) |
| 0.1 | 0.5 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 1.0 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.5 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.7 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.5 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 3.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.7 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.5 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.3 | 2.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 1.0 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.2 | 0.7 | GO:0004411 | homogentisate 1,2-dioxygenase activity(GO:0004411) |
| 0.2 | 0.8 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.2 | 2.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 1.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.2 | 1.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 1.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 3.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.5 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.6 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.6 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 1.0 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 1.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.3 | GO:0008513 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 0.2 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 2.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.8 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.4 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 1.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.5 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.2 | GO:0038047 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.0 | 3.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.7 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.4 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.4 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0000252 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) sterol-4-alpha-carboxylate 3-dehydrogenase (decarboxylating) activity(GO:0047012) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 2.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 2.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 5.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 3.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 2.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 2.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 2.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |