Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
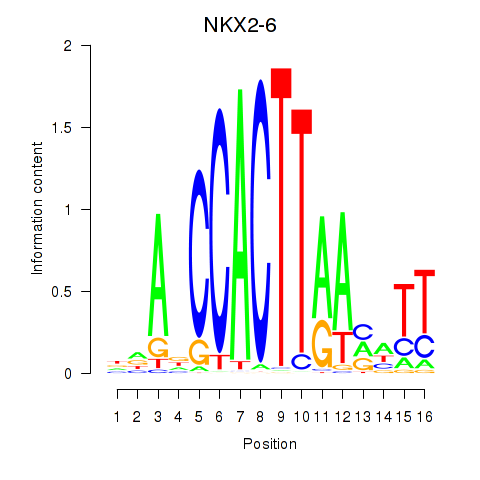
Results for NKX2-6
Z-value: 1.24
Transcription factors associated with NKX2-6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-6
|
ENSG00000180053.8 | NK2 homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-6 | hg38_v1_chr8_-_23706756_23706756 | -0.14 | 4.6e-01 | Click! |
Activity profile of NKX2-6 motif
Sorted Z-values of NKX2-6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_142791635 | 7.00 |
ENST00000633705.1
|
TRBC1
|
T cell receptor beta constant 1 |
| chr7_+_142800957 | 6.87 |
ENST00000466254.1
|
TRBC2
|
T cell receptor beta constant 2 |
| chr14_-_106579223 | 6.50 |
ENST00000390626.2
|
IGHV5-51
|
immunoglobulin heavy variable 5-51 |
| chr14_+_22320128 | 5.06 |
ENST00000390468.1
|
TRAV41
|
T cell receptor alpha variable 41 |
| chr19_+_3178756 | 4.32 |
ENST00000246115.5
|
S1PR4
|
sphingosine-1-phosphate receptor 4 |
| chr22_+_22887780 | 4.29 |
ENST00000532223.2
ENST00000526893.6 ENST00000531372.1 |
IGLL5
|
immunoglobulin lambda like polypeptide 5 |
| chr6_-_159044980 | 4.26 |
ENST00000367066.8
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr6_-_159045104 | 4.22 |
ENST00000326965.7
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr22_-_37486357 | 4.20 |
ENST00000356998.8
ENST00000416983.7 ENST00000424765.2 |
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr6_-_159045010 | 4.08 |
ENST00000338313.5
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr17_-_31318818 | 4.06 |
ENST00000578584.5
|
ENSG00000265118.5
|
novel protein |
| chr14_-_106762576 | 3.99 |
ENST00000624687.1
|
IGHV1-69D
|
immunoglobulin heavy variable 1-69D |
| chr3_-_121660892 | 3.94 |
ENST00000428394.6
ENST00000314583.8 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr11_-_111379268 | 3.91 |
ENST00000393067.8
|
POU2AF1
|
POU class 2 homeobox associating factor 1 |
| chr22_-_23754376 | 3.87 |
ENST00000398465.3
ENST00000248948.4 |
VPREB3
|
V-set pre-B cell surrogate light chain 3 |
| chr22_+_44181732 | 3.87 |
ENST00000415224.5
|
PARVG
|
parvin gamma |
| chr14_-_59870752 | 3.82 |
ENST00000611068.1
ENST00000267484.10 |
RTN1
|
reticulon 1 |
| chr12_-_14950606 | 3.62 |
ENST00000536592.5
|
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr6_+_32439866 | 3.43 |
ENST00000374982.5
ENST00000395388.7 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr3_+_114294020 | 3.38 |
ENST00000383671.8
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr14_-_105940235 | 3.04 |
ENST00000390593.2
|
IGHV6-1
|
immunoglobulin heavy variable 6-1 |
| chr15_+_81182579 | 2.95 |
ENST00000302987.9
|
IL16
|
interleukin 16 |
| chr14_-_106108453 | 2.81 |
ENST00000632950.2
|
IGHV5-10-1
|
immunoglobulin heavy variable 5-10-1 |
| chr2_-_181657096 | 2.78 |
ENST00000410087.8
ENST00000409440.7 |
CERKL
|
ceramide kinase like |
| chr6_-_33009568 | 2.75 |
ENST00000374813.1
ENST00000229829.7 |
HLA-DOA
|
major histocompatibility complex, class II, DO alpha |
| chr1_-_169703329 | 2.74 |
ENST00000497295.1
|
SELL
|
selectin L |
| chr3_+_186996444 | 2.51 |
ENST00000676633.1
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr22_+_44181357 | 2.38 |
ENST00000417767.1
|
PARVG
|
parvin gamma |
| chr17_+_4932248 | 2.34 |
ENST00000329125.6
|
GP1BA
|
glycoprotein Ib platelet subunit alpha |
| chr14_-_106211453 | 2.33 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr1_+_158289916 | 2.33 |
ENST00000368170.8
|
CD1C
|
CD1c molecule |
| chr17_+_4932285 | 2.19 |
ENST00000611961.1
|
GP1BA
|
glycoprotein Ib platelet subunit alpha |
| chr15_-_55270874 | 2.14 |
ENST00000567380.5
ENST00000565972.5 ENST00000569493.5 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr2_-_174597728 | 2.12 |
ENST00000409891.5
ENST00000410117.5 |
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr2_-_174597795 | 2.05 |
ENST00000679041.1
|
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr4_-_48080172 | 2.04 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr11_+_1870252 | 2.01 |
ENST00000612798.4
|
LSP1
|
lymphocyte specific protein 1 |
| chr2_+_69915041 | 2.00 |
ENST00000540449.5
|
MXD1
|
MAX dimerization protein 1 |
| chr1_+_109619827 | 1.99 |
ENST00000667949.2
ENST00000342115.8 ENST00000528667.7 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr3_-_46208304 | 1.99 |
ENST00000296140.4
|
CCR1
|
C-C motif chemokine receptor 1 |
| chr17_-_59151794 | 1.98 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr2_+_69915100 | 1.94 |
ENST00000264444.7
|
MXD1
|
MAX dimerization protein 1 |
| chr11_-_65862026 | 1.92 |
ENST00000532134.5
|
CFL1
|
cofilin 1 |
| chr2_-_174598206 | 1.90 |
ENST00000392546.6
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr8_+_27311620 | 1.89 |
ENST00000522338.5
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr22_+_44181844 | 1.86 |
ENST00000466375.2
|
PARVG
|
parvin gamma |
| chr22_+_22395005 | 1.83 |
ENST00000390298.2
|
IGLV7-43
|
immunoglobulin lambda variable 7-43 |
| chr12_-_75209701 | 1.83 |
ENST00000350228.6
ENST00000298972.5 |
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr17_+_59729702 | 1.83 |
ENST00000587259.5
|
VMP1
|
vacuole membrane protein 1 |
| chr11_-_62754141 | 1.82 |
ENST00000527994.1
ENST00000394807.5 ENST00000673933.1 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr11_-_65857007 | 1.80 |
ENST00000527344.5
|
CFL1
|
cofilin 1 |
| chr8_+_55102012 | 1.75 |
ENST00000327381.7
|
XKR4
|
XK related 4 |
| chr14_+_61485391 | 1.75 |
ENST00000640011.1
|
PRKCH
|
protein kinase C eta |
| chr11_-_65856944 | 1.72 |
ENST00000524553.5
|
CFL1
|
cofilin 1 |
| chr1_+_158930778 | 1.66 |
ENST00000458222.5
|
PYHIN1
|
pyrin and HIN domain family member 1 |
| chr3_+_122055355 | 1.66 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chr19_+_54874218 | 1.64 |
ENST00000355524.8
ENST00000391726.7 |
FCAR
|
Fc fragment of IgA receptor |
| chr22_-_50085378 | 1.63 |
ENST00000442311.1
|
MLC1
|
modulator of VRAC current 1 |
| chr7_+_150801695 | 1.63 |
ENST00000475536.5
ENST00000468689.2 |
TMEM176A
|
transmembrane protein 176A |
| chr2_-_100105339 | 1.61 |
ENST00000424600.5
ENST00000441400.5 ENST00000673328.1 |
AFF3
|
AF4/FMR2 family member 3 |
| chr14_+_21797272 | 1.61 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1 |
| chr1_+_158254414 | 1.59 |
ENST00000289429.6
|
CD1A
|
CD1a molecule |
| chr3_+_122077850 | 1.57 |
ENST00000482356.5
ENST00000393627.6 |
CD86
|
CD86 molecule |
| chr13_+_48653921 | 1.57 |
ENST00000682523.1
|
CYSLTR2
|
cysteinyl leukotriene receptor 2 |
| chr12_+_14408482 | 1.56 |
ENST00000541056.1
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr11_+_122838492 | 1.56 |
ENST00000227348.9
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr2_-_100104530 | 1.53 |
ENST00000432037.5
ENST00000673232.1 ENST00000423966.6 ENST00000409236.6 |
AFF3
|
AF4/FMR2 family member 3 |
| chr1_-_107688492 | 1.53 |
ENST00000415432.6
|
VAV3
|
vav guanine nucleotide exchange factor 3 |
| chr16_+_8712943 | 1.50 |
ENST00000561870.5
ENST00000396600.6 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr11_+_118883884 | 1.50 |
ENST00000292174.5
|
CXCR5
|
C-X-C motif chemokine receptor 5 |
| chr19_+_7637099 | 1.48 |
ENST00000595950.5
ENST00000221283.10 ENST00000441779.6 ENST00000414284.6 |
STXBP2
|
syntaxin binding protein 2 |
| chr3_+_122077776 | 1.46 |
ENST00000264468.9
|
CD86
|
CD86 molecule |
| chr19_+_54874275 | 1.42 |
ENST00000469767.5
ENST00000391725.7 ENST00000345937.8 ENST00000353758.8 ENST00000359272.8 ENST00000391723.7 ENST00000391724.3 |
FCAR
|
Fc fragment of IgA receptor |
| chr17_-_74531467 | 1.40 |
ENST00000314401.3
ENST00000392621.6 |
CD300LB
|
CD300 molecule like family member b |
| chr5_-_1523900 | 1.39 |
ENST00000283415.4
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1 |
| chr22_-_50085331 | 1.38 |
ENST00000395876.6
|
MLC1
|
modulator of VRAC current 1 |
| chr1_+_109620578 | 1.38 |
ENST00000531203.6
ENST00000256578.8 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr17_-_28576882 | 1.33 |
ENST00000395319.7
ENST00000581807.5 ENST00000226253.9 ENST00000584086.5 ENST00000395321.6 |
ALDOC
|
aldolase, fructose-bisphosphate C |
| chr19_-_43670153 | 1.31 |
ENST00000601723.5
ENST00000339082.7 ENST00000340093.8 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr11_-_6619353 | 1.31 |
ENST00000642892.1
ENST00000645620.1 ENST00000533371.6 ENST00000647152.1 ENST00000644810.1 ENST00000299427.12 ENST00000682424.1 ENST00000644218.1 ENST00000528657.2 ENST00000531754.2 |
TPP1
|
tripeptidyl peptidase 1 |
| chr3_-_56916379 | 1.25 |
ENST00000496106.5
|
ARHGEF3
|
Rho guanine nucleotide exchange factor 3 |
| chr6_+_31571957 | 1.25 |
ENST00000454783.5
|
LTA
|
lymphotoxin alpha |
| chr13_+_48653711 | 1.21 |
ENST00000614739.4
ENST00000617562.4 ENST00000621321.1 ENST00000622559.4 |
CYSLTR2
|
cysteinyl leukotriene receptor 2 |
| chr22_-_50085414 | 1.13 |
ENST00000311597.10
|
MLC1
|
modulator of VRAC current 1 |
| chr14_+_71321379 | 1.13 |
ENST00000557151.5
|
SIPA1L1
|
signal induced proliferation associated 1 like 1 |
| chr9_+_122370523 | 1.13 |
ENST00000643810.1
ENST00000540753.6 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr17_+_42998779 | 1.12 |
ENST00000586277.5
|
RPL27
|
ribosomal protein L27 |
| chr1_-_20786610 | 1.09 |
ENST00000375000.5
ENST00000312239.10 ENST00000419490.5 ENST00000414993.1 ENST00000443615.1 |
HP1BP3
|
heterochromatin protein 1 binding protein 3 |
| chr9_+_122371036 | 1.08 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr8_+_41529242 | 1.04 |
ENST00000523277.6
ENST00000276533.4 |
GINS4
|
GINS complex subunit 4 |
| chr16_+_8712831 | 1.04 |
ENST00000568847.5
|
ABAT
|
4-aminobutyrate aminotransferase |
| chr9_+_122371014 | 1.04 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr12_-_89526011 | 1.02 |
ENST00000313546.8
|
POC1B
|
POC1 centriolar protein B |
| chr12_-_89526164 | 1.01 |
ENST00000548729.5
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr4_-_46388713 | 0.98 |
ENST00000507069.5
|
GABRA2
|
gamma-aminobutyric acid type A receptor subunit alpha2 |
| chr8_-_144012748 | 0.96 |
ENST00000530478.5
|
PARP10
|
poly(ADP-ribose) polymerase family member 10 |
| chr1_-_1832282 | 0.95 |
ENST00000437146.1
|
GNB1
|
G protein subunit beta 1 |
| chr2_+_17541157 | 0.94 |
ENST00000406397.1
|
VSNL1
|
visinin like 1 |
| chr5_-_111756245 | 0.93 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr17_+_42998264 | 0.92 |
ENST00000589037.5
|
RPL27
|
ribosomal protein L27 |
| chr8_-_100950549 | 0.90 |
ENST00000395951.7
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta |
| chr4_-_46124046 | 0.89 |
ENST00000295452.5
|
GABRG1
|
gamma-aminobutyric acid type A receptor subunit gamma1 |
| chr1_-_77979054 | 0.87 |
ENST00000370768.7
ENST00000370767.5 ENST00000421641.1 |
FUBP1
|
far upstream element binding protein 1 |
| chr1_-_31065671 | 0.84 |
ENST00000440538.6
ENST00000424085.6 ENST00000257075.9 ENST00000373747.7 ENST00000426105.7 ENST00000525843.5 ENST00000373742.6 |
PUM1
|
pumilio RNA binding family member 1 |
| chr7_+_31687208 | 0.83 |
ENST00000409146.3
ENST00000342032.8 |
PPP1R17
|
protein phosphatase 1 regulatory subunit 17 |
| chr2_-_201433545 | 0.83 |
ENST00000440597.1
|
TRAK2
|
trafficking kinesin protein 2 |
| chr1_+_16440700 | 0.83 |
ENST00000504551.6
ENST00000457722.6 ENST00000337132.10 ENST00000443980.6 |
NECAP2
|
NECAP endocytosis associated 2 |
| chr11_-_56614874 | 0.82 |
ENST00000641076.1
|
OR5M1
|
olfactory receptor family 5 subfamily M member 1 |
| chr17_-_58529277 | 0.81 |
ENST00000579371.5
|
SEPTIN4
|
septin 4 |
| chr2_-_69643152 | 0.80 |
ENST00000606389.7
|
AAK1
|
AP2 associated kinase 1 |
| chrX_-_153321759 | 0.80 |
ENST00000436629.3
|
PNMA6F
|
PNMA family member 6F |
| chr19_-_2282168 | 0.79 |
ENST00000342063.5
|
PEAK3
|
PEAK family member 3 |
| chr6_+_28267107 | 0.79 |
ENST00000621053.1
ENST00000617168.4 ENST00000421553.7 ENST00000611552.2 ENST00000623276.3 |
ENSG00000276302.1
ZSCAN26
|
novel protein zinc finger and SCAN domain containing 26 |
| chr4_+_17577487 | 0.78 |
ENST00000606142.5
|
LAP3
|
leucine aminopeptidase 3 |
| chr17_-_51260032 | 0.78 |
ENST00000586178.6
|
MBTD1
|
mbt domain containing 1 |
| chr12_-_89526253 | 0.78 |
ENST00000547474.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr2_+_102311502 | 0.77 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr12_-_262828 | 0.76 |
ENST00000343164.9
ENST00000436453.1 ENST00000445055.6 ENST00000546319.5 |
SLC6A13
|
solute carrier family 6 member 13 |
| chr12_+_118016690 | 0.75 |
ENST00000537315.5
ENST00000454402.7 ENST00000484086.6 ENST00000420967.5 ENST00000392542.6 ENST00000535092.1 |
RFC5
|
replication factor C subunit 5 |
| chr6_-_89315291 | 0.75 |
ENST00000402938.4
|
GABRR2
|
gamma-aminobutyric acid type A receptor subunit rho2 |
| chr1_+_158999963 | 0.74 |
ENST00000566111.5
|
IFI16
|
interferon gamma inducible protein 16 |
| chrX_-_53281609 | 0.73 |
ENST00000638630.1
ENST00000375365.2 |
IQSEC2
|
IQ motif and Sec7 domain ArfGEF 2 |
| chr7_+_121873089 | 0.72 |
ENST00000651065.1
|
PTPRZ1
|
protein tyrosine phosphatase receptor type Z1 |
| chr2_-_181656974 | 0.72 |
ENST00000374969.6
ENST00000339098.9 ENST00000374970.6 |
CERKL
|
ceramide kinase like |
| chr20_-_1393074 | 0.71 |
ENST00000614856.2
ENST00000678408.1 ENST00000618612.5 ENST00000439640.5 ENST00000381719.8 ENST00000677533.1 |
FKBP1A
|
FKBP prolyl isomerase 1A |
| chr11_-_790062 | 0.69 |
ENST00000330106.5
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr20_-_1393045 | 0.68 |
ENST00000400137.9
ENST00000381715.4 |
FKBP1A
|
FKBP prolyl isomerase 1A |
| chr12_+_59689337 | 0.66 |
ENST00000261187.8
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr17_-_58529303 | 0.65 |
ENST00000580844.5
|
SEPTIN4
|
septin 4 |
| chr11_+_63369779 | 0.65 |
ENST00000279178.4
|
SLC22A9
|
solute carrier family 22 member 9 |
| chr17_-_58529344 | 0.64 |
ENST00000317268.7
|
SEPTIN4
|
septin 4 |
| chrX_-_47659128 | 0.62 |
ENST00000333119.7
ENST00000335890.3 |
UXT
|
ubiquitously expressed prefoldin like chaperone |
| chr9_+_124261427 | 0.61 |
ENST00000373596.5
ENST00000425237.5 |
NEK6
|
NIMA related kinase 6 |
| chrX_-_154409278 | 0.60 |
ENST00000369808.7
|
DNASE1L1
|
deoxyribonuclease 1 like 1 |
| chr21_-_29939532 | 0.57 |
ENST00000327783.8
ENST00000389124.6 ENST00000389125.7 ENST00000399913.5 |
GRIK1
|
glutamate ionotropic receptor kainate type subunit 1 |
| chr14_-_106875069 | 0.57 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr21_-_44230089 | 0.57 |
ENST00000643466.1
|
ICOSLG
|
inducible T cell costimulator ligand |
| chr6_+_28267044 | 0.57 |
ENST00000316606.10
|
ZSCAN26
|
zinc finger and SCAN domain containing 26 |
| chrX_-_47482529 | 0.55 |
ENST00000313116.11
|
ZNF41
|
zinc finger protein 41 |
| chr12_+_32106762 | 0.54 |
ENST00000551848.1
ENST00000652176.1 |
BICD1
|
BICD cargo adaptor 1 |
| chr1_-_149813687 | 0.53 |
ENST00000331491.2
|
H3C13
|
H3 clustered histone 13 |
| chr2_+_115064660 | 0.53 |
ENST00000393146.6
|
DPP10
|
dipeptidyl peptidase like 10 |
| chr17_+_42998379 | 0.52 |
ENST00000253788.12
ENST00000589913.6 |
RPL27
|
ribosomal protein L27 |
| chr19_-_51108365 | 0.52 |
ENST00000421832.3
|
CTU1
|
cytosolic thiouridylase subunit 1 |
| chr4_+_168497113 | 0.50 |
ENST00000511948.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr7_-_91880672 | 0.49 |
ENST00000456229.1
ENST00000442961.1 ENST00000406735.6 ENST00000419292.1 ENST00000351870.8 |
MTERF1
|
mitochondrial transcription termination factor 1 |
| chr12_+_54280842 | 0.49 |
ENST00000678077.1
ENST00000548688.5 |
HNRNPA1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr12_-_43758748 | 0.48 |
ENST00000416848.6
ENST00000550784.5 ENST00000547156.1 ENST00000549868.1 ENST00000551923.5 ENST00000344862.10 ENST00000431332.7 |
PUS7L
|
pseudouridine synthase 7 like |
| chr3_+_171843337 | 0.48 |
ENST00000334567.9
ENST00000619900.4 ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chrY_-_23694579 | 0.48 |
ENST00000343584.10
|
PRYP3
|
PTPN13 like Y-linked pseudogene 3 |
| chr12_+_54280663 | 0.48 |
ENST00000677375.1
ENST00000677210.1 ENST00000677385.1 ENST00000677249.1 ENST00000550482.2 ENST00000679101.1 ENST00000340913.11 ENST00000547708.5 ENST00000551702.5 ENST00000676794.1 ENST00000330752.12 ENST00000678690.1 ENST00000678919.1 ENST00000547276.5 |
HNRNPA1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr11_-_129947576 | 0.48 |
ENST00000423662.6
|
PRDM10
|
PR/SET domain 10 |
| chr4_-_170003738 | 0.47 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibril associated protein 3 like |
| chr7_+_141764097 | 0.47 |
ENST00000247879.2
|
TAS2R3
|
taste 2 receptor member 3 |
| chr1_-_120051714 | 0.47 |
ENST00000579475.7
|
NOTCH2
|
notch receptor 2 |
| chrX_-_154409246 | 0.46 |
ENST00000369807.6
|
DNASE1L1
|
deoxyribonuclease 1 like 1 |
| chr12_+_43758936 | 0.44 |
ENST00000440781.6
ENST00000431837.5 ENST00000550616.5 ENST00000613694.5 ENST00000551736.5 |
IRAK4
|
interleukin 1 receptor associated kinase 4 |
| chr6_+_125781108 | 0.44 |
ENST00000368357.7
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr9_-_101738640 | 0.43 |
ENST00000361820.6
|
GRIN3A
|
glutamate ionotropic receptor NMDA type subunit 3A |
| chr8_+_22066489 | 0.43 |
ENST00000522340.5
ENST00000519333.6 |
DMTN
|
dematin actin binding protein |
| chr22_+_29205877 | 0.43 |
ENST00000334018.11
ENST00000429226.5 ENST00000404755.7 ENST00000404820.7 ENST00000430127.1 |
EMID1
|
EMI domain containing 1 |
| chr8_-_133258387 | 0.42 |
ENST00000676222.1
|
NDRG1
|
N-myc downstream regulated 1 |
| chr12_-_75209422 | 0.42 |
ENST00000393288.2
ENST00000540018.5 |
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr21_-_45544392 | 0.42 |
ENST00000443742.1
ENST00000528477.1 ENST00000567670.5 |
SLC19A1
|
solute carrier family 19 member 1 |
| chr1_-_173824322 | 0.42 |
ENST00000356198.6
|
CENPL
|
centromere protein L |
| chr1_+_159826860 | 0.41 |
ENST00000289707.10
|
SLAMF8
|
SLAM family member 8 |
| chr5_-_134174765 | 0.40 |
ENST00000520417.1
|
SKP1
|
S-phase kinase associated protein 1 |
| chr7_-_30682473 | 0.40 |
ENST00000506074.6
|
CRHR2
|
corticotropin releasing hormone receptor 2 |
| chr7_+_70766172 | 0.40 |
ENST00000644949.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr12_-_7936177 | 0.40 |
ENST00000544291.1
ENST00000075120.12 |
SLC2A3
|
solute carrier family 2 member 3 |
| chr1_-_70354673 | 0.39 |
ENST00000370944.9
ENST00000262346.6 |
ANKRD13C
|
ankyrin repeat domain 13C |
| chr11_+_4903783 | 0.39 |
ENST00000641490.1
|
OR51A7
|
olfactory receptor family 51 subfamily A member 7 |
| chr12_-_38906269 | 0.38 |
ENST00000550863.1
|
CPNE8
|
copine 8 |
| chr2_-_27323072 | 0.38 |
ENST00000428910.5
ENST00000402722.5 ENST00000380044.6 ENST00000399052.8 ENST00000405076.5 |
MPV17
|
mitochondrial inner membrane protein MPV17 |
| chr7_+_121873152 | 0.38 |
ENST00000650826.1
ENST00000650728.1 ENST00000393386.7 ENST00000651390.1 ENST00000651842.1 ENST00000650681.1 |
PTPRZ1
|
protein tyrosine phosphatase receptor type Z1 |
| chr1_-_173824107 | 0.36 |
ENST00000345664.10
ENST00000367710.7 |
CENPL
|
centromere protein L |
| chr5_-_131994225 | 0.36 |
ENST00000543479.5
ENST00000431707.5 |
ACSL6
|
acyl-CoA synthetase long chain family member 6 |
| chr18_+_13443720 | 0.35 |
ENST00000678309.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr11_+_65860394 | 0.35 |
ENST00000308110.9
|
MUS81
|
MUS81 structure-specific endonuclease subunit |
| chr1_-_109613070 | 0.35 |
ENST00000351050.8
|
GNAT2
|
G protein subunit alpha transducin 2 |
| chr9_-_19065084 | 0.35 |
ENST00000415524.1
|
HAUS6
|
HAUS augmin like complex subunit 6 |
| chr19_-_12597298 | 0.35 |
ENST00000440366.1
|
ZNF490
|
zinc finger protein 490 |
| chr8_+_84184875 | 0.35 |
ENST00000517638.5
ENST00000522647.1 |
RALYL
|
RALY RNA binding protein like |
| chr15_-_65286837 | 0.34 |
ENST00000444347.2
ENST00000261888.10 ENST00000649807.2 |
PARP16
|
poly(ADP-ribose) polymerase family member 16 |
| chr15_+_67067780 | 0.34 |
ENST00000679624.1
|
SMAD3
|
SMAD family member 3 |
| chr13_+_27424583 | 0.34 |
ENST00000381140.10
|
GTF3A
|
general transcription factor IIIA |
| chr17_-_36534927 | 0.33 |
ENST00000610930.4
|
MYO19
|
myosin XIX |
| chr19_+_51571249 | 0.33 |
ENST00000262259.7
ENST00000545217.5 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr1_+_205256189 | 0.33 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr1_+_28369705 | 0.32 |
ENST00000373839.8
|
PHACTR4
|
phosphatase and actin regulator 4 |
| chr3_-_39154558 | 0.32 |
ENST00000514182.1
|
CSRNP1
|
cysteine and serine rich nuclear protein 1 |
| chr10_+_119892692 | 0.32 |
ENST00000369075.8
|
SEC23IP
|
SEC23 interacting protein |
| chr17_+_68515082 | 0.32 |
ENST00000588178.5
|
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chr7_-_6826275 | 0.32 |
ENST00000316731.13
|
CCZ1B
|
CCZ1 homolog B, vacuolar protein trafficking and biogenesis associated |
| chr20_+_56392607 | 0.31 |
ENST00000217109.9
ENST00000452950.1 |
CSTF1
|
cleavage stimulation factor subunit 1 |
| chr17_-_3063607 | 0.31 |
ENST00000575751.1
|
OR1D5
|
olfactory receptor family 1 subfamily D member 5 |
| chr10_-_5977589 | 0.30 |
ENST00000620345.4
ENST00000397251.7 ENST00000397248.6 ENST00000622442.4 ENST00000620865.4 |
IL15RA
|
interleukin 15 receptor subunit alpha |
| chr6_+_28267355 | 0.30 |
ENST00000614088.1
ENST00000619937.4 |
ZSCAN26
|
zinc finger and SCAN domain containing 26 |
| chr1_+_173824956 | 0.30 |
ENST00000648458.1
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr17_+_35587239 | 0.29 |
ENST00000621914.4
ENST00000621668.4 ENST00000616681.4 ENST00000612035.4 ENST00000610402.5 ENST00000614600.4 ENST00000590432.5 ENST00000612116.5 |
AP2B1
|
adaptor related protein complex 2 subunit beta 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0002665 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 1.1 | 3.4 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.9 | 2.7 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.8 | 5.4 | GO:2000814 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.6 | 3.6 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.6 | 2.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.4 | 1.2 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.4 | 34.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.4 | 1.9 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.4 | 2.5 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.4 | 3.9 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.3 | 2.8 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.3 | 3.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 2.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.3 | 2.0 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.3 | 1.4 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.3 | 4.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.3 | 1.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.2 | 3.9 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 0.6 | GO:0070145 | mitochondrial asparaginyl-tRNA aminoacylation(GO:0070145) |
| 0.2 | 4.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.2 | 1.3 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.2 | 1.6 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 4.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.2 | 0.7 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.2 | 3.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 4.5 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 1.8 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 0.8 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.1 | 1.0 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.1 | 0.5 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 3.4 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.1 | 1.6 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.1 | 2.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.8 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.1 | 0.7 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.1 | 1.6 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.1 | 0.3 | GO:1904226 | regulation of UDP-glucose catabolic process(GO:0010904) negative regulation of UDP-glucose catabolic process(GO:0010905) regulation of glycogen synthase activity, transferring glucose-1-phosphate(GO:1904226) negative regulation of glycogen synthase activity, transferring glucose-1-phosphate(GO:1904227) |
| 0.1 | 0.7 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 2.5 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.5 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.4 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 1.4 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.1 | 2.0 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 0.8 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 0.4 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 1.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.7 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 1.1 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 1.5 | GO:0048535 | lymph node development(GO:0048535) |
| 0.1 | 0.9 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 1.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 2.7 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 2.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 8.6 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 8.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.9 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 2.5 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.8 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.4 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 | 1.1 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.4 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 1.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.4 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.5 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.9 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.6 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.0 | 1.7 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 13.9 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.8 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.6 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 1.0 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 1.1 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 1.3 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.1 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.0 | 1.0 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.8 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 2.0 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.5 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.6 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.4 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.6 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 2.9 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.7 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.0 | 0.3 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.3 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 2.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 2.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.5 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 1.0 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 34.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.4 | 2.5 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.3 | 6.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 1.4 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.3 | 1.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 1.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 1.5 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 4.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.5 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 2.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 2.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.6 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 4.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 0.4 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 4.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 2.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 2.2 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 3.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 10.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 3.5 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.1 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 2.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 3.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.0 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 2.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 3.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 3.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network(GO:0031094) platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 4.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.1 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 7.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.8 | 3.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.6 | 3.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.5 | 4.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.5 | 34.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 3.9 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.4 | 1.9 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.4 | 2.5 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.4 | 6.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.4 | 3.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 2.8 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.3 | 1.4 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.3 | 2.7 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.3 | 3.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 4.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.3 | 3.5 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.3 | 0.8 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.3 | 2.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 1.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.2 | 5.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.3 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.2 | 1.3 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.2 | 0.6 | GO:0050560 | aspartate-tRNA ligase activity(GO:0004815) aspartate-tRNA(Asn) ligase activity(GO:0050560) |
| 0.2 | 1.8 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 0.6 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.2 | 0.7 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 1.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 2.0 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 2.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 1.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.1 | 1.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.3 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.3 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 1.0 | GO:0060002 | myosin light chain binding(GO:0032027) plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.4 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.5 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.7 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 4.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.4 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 2.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 2.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.0 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.6 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.6 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 3.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.9 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.5 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.0 | 5.4 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 4.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 3.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 12.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.0 | 4.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 4.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 1.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 8.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 4.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 5.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 0.9 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 9.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 4.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 2.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 7.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 4.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 4.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 5.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 4.7 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 4.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 3.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.8 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 2.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 18.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.8 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 2.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 4.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.7 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 1.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 3.5 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 2.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.6 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 2.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |