Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
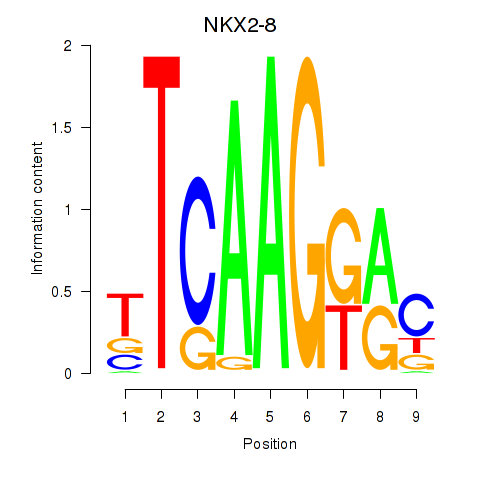
Results for NKX2-8
Z-value: 1.13
Transcription factors associated with NKX2-8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-8
|
ENSG00000136327.7 | NK2 homeobox 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-8 | hg38_v1_chr14_-_36582593_36582626 | -0.00 | 9.9e-01 | Click! |
Activity profile of NKX2-8 motif
Sorted Z-values of NKX2-8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_107719657 | 4.21 |
ENST00000525934.1
ENST00000531293.1 |
SLN
|
sarcolipin |
| chr14_-_106235582 | 2.72 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr12_+_13196718 | 2.51 |
ENST00000431267.2
ENST00000542474.5 ENST00000544053.5 ENST00000256951.10 |
EMP1
|
epithelial membrane protein 1 |
| chr14_-_106675544 | 2.46 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr11_-_19201976 | 2.34 |
ENST00000647990.1
ENST00000649235.1 ENST00000265968.9 ENST00000649842.1 |
CSRP3
|
cysteine and glycine rich protein 3 |
| chr14_-_106130061 | 2.34 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr14_-_106593319 | 2.31 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr14_-_106335613 | 2.27 |
ENST00000603660.1
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr12_+_13196777 | 2.14 |
ENST00000538364.5
ENST00000396301.7 |
EMP1
|
epithelial membrane protein 1 |
| chr2_-_89010515 | 2.06 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr3_-_52454032 | 2.03 |
ENST00000232975.8
|
TNNC1
|
troponin C1, slow skeletal and cardiac type |
| chr14_-_106269133 | 1.90 |
ENST00000390609.3
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr14_-_106538331 | 1.87 |
ENST00000390624.3
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr14_-_105626066 | 1.83 |
ENST00000641978.1
ENST00000390543.3 |
IGHG4
|
immunoglobulin heavy constant gamma 4 (G4m marker) |
| chr16_-_33845229 | 1.81 |
ENST00000569103.2
|
IGHV3OR16-17
|
immunoglobulin heavy variable 3/OR16-17 (non-functional) |
| chr14_-_106579223 | 1.81 |
ENST00000390626.2
|
IGHV5-51
|
immunoglobulin heavy variable 5-51 |
| chr14_-_106211453 | 1.79 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr14_-_106360320 | 1.74 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
| chr14_-_106811131 | 1.73 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr5_+_81233314 | 1.67 |
ENST00000511719.5
ENST00000437669.5 ENST00000254035.9 ENST00000424301.6 ENST00000505060.1 |
CKMT2
|
creatine kinase, mitochondrial 2 |
| chr14_-_106088573 | 1.60 |
ENST00000632099.1
|
IGHV3-64D
|
immunoglobulin heavy variable 3-64D |
| chr14_-_106791226 | 1.55 |
ENST00000433072.2
|
IGHV3-72
|
immunoglobulin heavy variable 3-72 |
| chr10_-_73641450 | 1.53 |
ENST00000359322.5
|
MYOZ1
|
myozenin 1 |
| chr16_+_32995048 | 1.52 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr6_+_96563117 | 1.41 |
ENST00000450218.6
|
FHL5
|
four and a half LIM domains 5 |
| chr14_-_106389858 | 1.40 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr14_-_106507476 | 1.32 |
ENST00000390621.3
|
IGHV1-45
|
immunoglobulin heavy variable 1-45 |
| chr15_+_51681483 | 1.23 |
ENST00000542355.6
ENST00000220478.8 ENST00000558709.1 |
SCG3
|
secretogranin III |
| chr14_-_106875069 | 1.22 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr11_-_19202004 | 1.17 |
ENST00000648719.1
|
CSRP3
|
cysteine and glycine rich protein 3 |
| chr16_+_33827140 | 1.11 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr14_-_106470788 | 1.10 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr14_-_105856183 | 1.07 |
ENST00000637539.2
ENST00000390559.6 |
IGHM
|
immunoglobulin heavy constant mu |
| chr4_-_175787382 | 1.05 |
ENST00000507520.5
|
GPM6A
|
glycoprotein M6A |
| chr6_-_131951364 | 1.02 |
ENST00000367976.4
|
CCN2
|
cellular communication network factor 2 |
| chr17_-_3691887 | 1.01 |
ENST00000552050.5
|
P2RX5
|
purinergic receptor P2X 5 |
| chr10_-_15719885 | 1.00 |
ENST00000378076.4
|
ITGA8
|
integrin subunit alpha 8 |
| chr17_-_44911281 | 0.91 |
ENST00000638304.1
ENST00000591880.2 ENST00000586125.2 ENST00000639921.1 |
GFAP
|
glial fibrillary acidic protein |
| chr3_+_111998739 | 0.90 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr12_-_85836372 | 0.89 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9 |
| chr17_-_15260752 | 0.88 |
ENST00000676329.1
ENST00000675551.1 ENST00000644020.1 ENST00000674947.1 |
PMP22
|
peripheral myelin protein 22 |
| chr6_-_32763466 | 0.87 |
ENST00000427449.1
ENST00000411527.5 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr5_-_79512794 | 0.86 |
ENST00000282260.10
ENST00000508576.5 ENST00000535690.1 |
HOMER1
|
homer scaffold protein 1 |
| chrX_+_103774385 | 0.85 |
ENST00000494475.5
|
PLP1
|
proteolipid protein 1 |
| chr19_+_48469354 | 0.85 |
ENST00000452733.7
ENST00000641098.1 |
CYTH2
|
cytohesin 2 |
| chr11_-_72642450 | 0.84 |
ENST00000444035.6
ENST00000544570.5 |
PDE2A
|
phosphodiesterase 2A |
| chr14_+_63204436 | 0.76 |
ENST00000316754.8
|
RHOJ
|
ras homolog family member J |
| chr18_-_58629084 | 0.74 |
ENST00000361673.4
|
ALPK2
|
alpha kinase 2 |
| chr11_-_72642407 | 0.74 |
ENST00000376450.7
|
PDE2A
|
phosphodiesterase 2A |
| chr4_-_46993520 | 0.73 |
ENST00000264318.4
|
GABRA4
|
gamma-aminobutyric acid type A receptor subunit alpha4 |
| chr18_+_45724127 | 0.73 |
ENST00000619403.4
ENST00000587601.5 |
SLC14A1
|
solute carrier family 14 member 1 (Kidd blood group) |
| chr3_+_111998915 | 0.72 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chrX_+_153687918 | 0.71 |
ENST00000253122.10
|
SLC6A8
|
solute carrier family 6 member 8 |
| chr16_+_172869 | 0.69 |
ENST00000251595.11
ENST00000397806.1 ENST00000484216.1 |
HBA2
|
hemoglobin subunit alpha 2 |
| chr19_-_4559663 | 0.69 |
ENST00000586582.6
|
SEMA6B
|
semaphorin 6B |
| chr22_+_22343185 | 0.66 |
ENST00000427632.2
|
IGLV9-49
|
immunoglobulin lambda variable 9-49 |
| chr19_-_48993300 | 0.66 |
ENST00000323798.8
ENST00000263276.6 |
GYS1
|
glycogen synthase 1 |
| chr5_+_141094595 | 0.66 |
ENST00000622947.1
ENST00000194155.7 ENST00000624874.1 ENST00000625033.1 |
PCDHB2
|
protocadherin beta 2 |
| chr2_-_63588390 | 0.64 |
ENST00000272321.12
ENST00000409562.7 |
WDPCP
|
WD repeat containing planar cell polarity effector |
| chr3_-_196515315 | 0.64 |
ENST00000397537.3
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr12_-_21657831 | 0.63 |
ENST00000450584.5
ENST00000350669.5 ENST00000673047.2 |
LDHB
|
lactate dehydrogenase B |
| chr5_-_176629943 | 0.63 |
ENST00000510387.5
ENST00000506696.1 |
SNCB
|
synuclein beta |
| chr14_+_63204859 | 0.63 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J |
| chr21_+_32298849 | 0.62 |
ENST00000303645.10
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr1_+_37807762 | 0.62 |
ENST00000373042.5
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr12_-_21657792 | 0.61 |
ENST00000396076.5
|
LDHB
|
lactate dehydrogenase B |
| chr16_+_28863812 | 0.60 |
ENST00000684370.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr8_+_66493556 | 0.59 |
ENST00000305454.8
ENST00000522977.5 ENST00000480005.1 |
VXN
|
vexin |
| chr16_+_176659 | 0.57 |
ENST00000320868.9
ENST00000397797.1 |
HBA1
|
hemoglobin subunit alpha 1 |
| chr9_+_35909481 | 0.56 |
ENST00000443779.3
|
SPAAR
|
small regulatory polypeptide of amino acid response |
| chr17_-_44968263 | 0.55 |
ENST00000253407.4
|
C1QL1
|
complement C1q like 1 |
| chr21_+_32298945 | 0.53 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr10_+_17752185 | 0.53 |
ENST00000377495.2
|
TMEM236
|
transmembrane protein 236 |
| chr16_+_22490337 | 0.53 |
ENST00000415833.6
|
NPIPB5
|
nuclear pore complex interacting protein family member B5 |
| chr19_+_48469306 | 0.53 |
ENST00000460595.5
|
CYTH2
|
cytohesin 2 |
| chr19_-_33225844 | 0.52 |
ENST00000253188.8
|
SLC7A10
|
solute carrier family 7 member 10 |
| chr6_-_56693294 | 0.52 |
ENST00000652573.1
|
DST
|
dystonin |
| chr10_-_13972355 | 0.52 |
ENST00000264546.10
|
FRMD4A
|
FERM domain containing 4A |
| chr6_+_34466059 | 0.51 |
ENST00000620693.4
ENST00000244458.7 ENST00000374043.6 |
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr8_-_65842051 | 0.50 |
ENST00000401827.8
|
PDE7A
|
phosphodiesterase 7A |
| chr3_+_66998297 | 0.50 |
ENST00000484414.5
ENST00000460576.5 ENST00000417314.2 |
KBTBD8
|
kelch repeat and BTB domain containing 8 |
| chr3_-_8769602 | 0.48 |
ENST00000316793.8
ENST00000431493.1 |
OXTR
|
oxytocin receptor |
| chr3_-_180679468 | 0.47 |
ENST00000651046.1
ENST00000476379.6 |
CCDC39
|
coiled-coil domain containing 39 |
| chr1_-_94541636 | 0.46 |
ENST00000370207.4
|
F3
|
coagulation factor III, tissue factor |
| chr2_+_232633551 | 0.46 |
ENST00000264059.8
|
EFHD1
|
EF-hand domain family member D1 |
| chr11_+_27040725 | 0.45 |
ENST00000529202.5
ENST00000263182.8 |
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr19_-_11236497 | 0.45 |
ENST00000587656.5
|
DOCK6
|
dedicator of cytokinesis 6 |
| chr1_-_109389886 | 0.43 |
ENST00000493736.5
|
SORT1
|
sortilin 1 |
| chr9_+_35910076 | 0.43 |
ENST00000636776.1
|
SPAAR
|
small regulatory polypeptide of amino acid response |
| chr17_+_78214186 | 0.43 |
ENST00000301633.8
ENST00000350051.8 ENST00000374948.6 ENST00000590449.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr8_+_66493514 | 0.42 |
ENST00000521495.5
|
VXN
|
vexin |
| chr2_-_68319887 | 0.42 |
ENST00000409862.1
ENST00000263655.4 |
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr12_+_50112167 | 0.41 |
ENST00000548468.2
ENST00000552815.1 ENST00000550654.1 ENST00000550487.6 ENST00000548985.1 |
ENSG00000272368.2
COX14
|
novel transcript, antisense to CERS5 cytochrome c oxidase assembly factor COX14 |
| chr4_+_71062642 | 0.41 |
ENST00000649996.1
|
SLC4A4
|
solute carrier family 4 member 4 |
| chr2_+_227616998 | 0.41 |
ENST00000641801.1
|
SCYGR4
|
small cysteine and glycine repeat containing 4 |
| chr17_-_69150062 | 0.40 |
ENST00000522787.5
ENST00000521538.5 |
ABCA10
|
ATP binding cassette subfamily A member 10 |
| chr12_+_101568350 | 0.40 |
ENST00000550514.5
|
MYBPC1
|
myosin binding protein C1 |
| chr5_+_157180816 | 0.39 |
ENST00000422843.8
|
ITK
|
IL2 inducible T cell kinase |
| chr8_-_61646807 | 0.39 |
ENST00000522919.5
|
ASPH
|
aspartate beta-hydroxylase |
| chr1_-_93681829 | 0.39 |
ENST00000260502.11
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr12_+_50112028 | 0.38 |
ENST00000317943.6
|
COX14
|
cytochrome c oxidase assembly factor COX14 |
| chr2_-_157628852 | 0.38 |
ENST00000243349.13
|
ACVR1C
|
activin A receptor type 1C |
| chr22_+_38468036 | 0.36 |
ENST00000409006.3
ENST00000216014.9 |
KDELR3
|
KDEL endoplasmic reticulum protein retention receptor 3 |
| chrX_-_151974668 | 0.36 |
ENST00000370328.4
|
GABRE
|
gamma-aminobutyric acid type A receptor subunit epsilon |
| chr8_-_61646863 | 0.36 |
ENST00000519678.5
|
ASPH
|
aspartate beta-hydroxylase |
| chr5_+_77086682 | 0.35 |
ENST00000643365.1
ENST00000645183.1 ENST00000645374.1 ENST00000647364.1 ENST00000643848.1 ENST00000643603.1 ENST00000645459.1 ENST00000643269.1 ENST00000503969.6 ENST00000646262.1 |
ZBED3-AS1
ENSG00000284762.1
|
ZBED3 antisense RNA 1 phosphodiesterase 8B |
| chr12_-_10167591 | 0.35 |
ENST00000543993.5
ENST00000339968.6 |
OLR1
|
oxidized low density lipoprotein receptor 1 |
| chr11_-_5249836 | 0.35 |
ENST00000632727.1
ENST00000330597.5 |
HBG1
|
hemoglobin subunit gamma 1 |
| chr7_-_100119291 | 0.34 |
ENST00000431404.2
|
TAF6
|
TATA-box binding protein associated factor 6 |
| chr9_-_14300231 | 0.34 |
ENST00000636735.1
|
NFIB
|
nuclear factor I B |
| chr8_+_124539097 | 0.33 |
ENST00000606244.2
ENST00000276689.8 ENST00000518008.5 ENST00000517367.1 |
NDUFB9
|
NADH:ubiquinone oxidoreductase subunit B9 |
| chr9_-_22009272 | 0.32 |
ENST00000380142.5
ENST00000276925.7 |
CDKN2B
|
cyclin dependent kinase inhibitor 2B |
| chrX_+_630789 | 0.31 |
ENST00000381575.6
|
SHOX
|
short stature homeobox |
| chr2_+_203936755 | 0.31 |
ENST00000316386.11
ENST00000435193.1 |
ICOS
|
inducible T cell costimulator |
| chr11_+_124012997 | 0.31 |
ENST00000641521.1
ENST00000641722.1 |
OR10G4
|
olfactory receptor family 10 subfamily G member 4 |
| chr4_+_140343443 | 0.31 |
ENST00000338517.8
ENST00000394203.7 ENST00000506322.5 |
SCOC
|
short coiled-coil protein |
| chr12_+_7789393 | 0.30 |
ENST00000229307.9
|
NANOG
|
Nanog homeobox |
| chr12_-_102197827 | 0.30 |
ENST00000329406.5
|
PMCH
|
pro-melanin concentrating hormone |
| chr12_-_75209814 | 0.29 |
ENST00000549446.6
|
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr11_-_17476801 | 0.29 |
ENST00000644772.1
ENST00000642271.1 ENST00000684571.1 ENST00000646902.1 ENST00000647015.1 ENST00000302539.9 ENST00000389817.8 ENST00000643260.1 ENST00000683136.1 |
ABCC8
|
ATP binding cassette subfamily C member 8 |
| chr1_+_75786246 | 0.29 |
ENST00000319942.8
|
RABGGTB
|
Rab geranylgeranyltransferase subunit beta |
| chr17_-_33293247 | 0.28 |
ENST00000225823.7
|
ASIC2
|
acid sensing ion channel subunit 2 |
| chr15_-_56465130 | 0.28 |
ENST00000260453.4
|
MNS1
|
meiosis specific nuclear structural 1 |
| chr4_-_122456725 | 0.28 |
ENST00000226730.5
|
IL2
|
interleukin 2 |
| chr17_+_78231310 | 0.27 |
ENST00000374946.7
ENST00000421688.5 ENST00000586400.5 |
TMEM235
|
transmembrane protein 235 |
| chr12_+_25052732 | 0.27 |
ENST00000547044.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr12_+_59664677 | 0.26 |
ENST00000548610.5
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr19_-_43754901 | 0.26 |
ENST00000270066.11
ENST00000601170.5 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr12_+_56996151 | 0.26 |
ENST00000556850.1
|
GPR182
|
G protein-coupled receptor 182 |
| chr17_-_7204502 | 0.26 |
ENST00000486626.8
ENST00000648263.1 |
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr11_-_40293102 | 0.25 |
ENST00000527150.5
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr7_+_142345412 | 0.25 |
ENST00000390392.3
|
TRBV4-2
|
T cell receptor beta variable 4-2 |
| chr13_-_51845169 | 0.25 |
ENST00000627246.3
ENST00000629372.3 |
TMEM272
|
transmembrane protein 272 |
| chr2_-_157628727 | 0.25 |
ENST00000335450.7
ENST00000348328.9 |
ACVR1C
|
activin A receptor type 1C |
| chr12_-_6493295 | 0.24 |
ENST00000540949.1
|
MRPL51
|
mitochondrial ribosomal protein L51 |
| chr11_+_75562056 | 0.24 |
ENST00000533603.5
|
SERPINH1
|
serpin family H member 1 |
| chr6_-_32666648 | 0.24 |
ENST00000399082.7
ENST00000399079.7 ENST00000374943.8 ENST00000434651.6 |
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr17_-_78782257 | 0.24 |
ENST00000591455.5
ENST00000446868.7 ENST00000361101.8 ENST00000589296.5 |
CYTH1
|
cytohesin 1 |
| chr3_+_173584433 | 0.24 |
ENST00000361589.8
|
NLGN1
|
neuroligin 1 |
| chr7_-_99466148 | 0.23 |
ENST00000394186.3
ENST00000359832.8 ENST00000449683.5 ENST00000292475.8 ENST00000488775.5 ENST00000523680.1 ENST00000430982.1 ENST00000413834.5 |
ATP5MF
PTCD1
ATP5MF-PTCD1
|
ATP synthase membrane subunit f pentatricopeptide repeat domain 1 ATP5MF-PTCD1 readthrough |
| chr5_-_181261078 | 0.23 |
ENST00000611618.1
|
TRIM52
|
tripartite motif containing 52 |
| chr14_+_24070837 | 0.23 |
ENST00000537691.5
ENST00000397016.6 ENST00000560356.5 ENST00000558450.5 |
CPNE6
|
copine 6 |
| chr5_+_43602648 | 0.23 |
ENST00000505678.6
ENST00000512422.5 ENST00000264663.9 ENST00000670904.1 ENST00000653251.1 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr8_+_103298836 | 0.23 |
ENST00000523739.5
ENST00000358755.5 |
FZD6
|
frizzled class receptor 6 |
| chr11_-_132943092 | 0.23 |
ENST00000612177.4
ENST00000541867.5 |
OPCML
|
opioid binding protein/cell adhesion molecule like |
| chr12_+_49265071 | 0.22 |
ENST00000549183.1
ENST00000639419.1 ENST00000301072.11 |
TUBA1C
|
tubulin alpha 1c |
| chr3_-_179266971 | 0.22 |
ENST00000349697.2
ENST00000497599.5 |
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr1_+_75786214 | 0.22 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase subunit beta |
| chrM_+_8489 | 0.22 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase membrane subunit 6 |
| chr8_+_97887903 | 0.22 |
ENST00000520016.5
|
MATN2
|
matrilin 2 |
| chr4_+_89894846 | 0.22 |
ENST00000264790.7
|
MMRN1
|
multimerin 1 |
| chr16_+_28863946 | 0.21 |
ENST00000395532.8
|
SH2B1
|
SH2B adaptor protein 1 |
| chrX_+_116436599 | 0.21 |
ENST00000598581.3
|
SLC6A14
|
solute carrier family 6 member 14 |
| chr16_+_58392391 | 0.20 |
ENST00000426538.6
ENST00000328514.11 |
GINS3
|
GINS complex subunit 3 |
| chr6_-_2744126 | 0.20 |
ENST00000647417.1
|
MYLK4
|
myosin light chain kinase family member 4 |
| chr22_+_29438583 | 0.20 |
ENST00000354373.2
|
RFPL1
|
ret finger protein like 1 |
| chr17_-_7208325 | 0.20 |
ENST00000650120.1
ENST00000648760.1 |
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr1_+_151060357 | 0.20 |
ENST00000368921.5
|
MLLT11
|
MLLT11 transcription factor 7 cofactor |
| chr17_+_78214286 | 0.20 |
ENST00000592734.5
ENST00000587746.5 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr12_+_25052512 | 0.20 |
ENST00000557489.5
ENST00000354454.7 ENST00000536173.5 |
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr12_-_10167657 | 0.20 |
ENST00000538745.5
|
OLR1
|
oxidized low density lipoprotein receptor 1 |
| chr7_+_100219236 | 0.19 |
ENST00000317271.2
|
PVRIG
|
PVR related immunoglobulin domain containing |
| chr1_+_32205657 | 0.19 |
ENST00000291358.11
ENST00000537469.2 |
IQCC
|
IQ motif containing C |
| chrX_-_48919015 | 0.19 |
ENST00000376509.4
|
PIM2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr5_+_43603163 | 0.19 |
ENST00000660752.1
ENST00000654405.1 ENST00000344920.9 ENST00000657172.1 ENST00000512996.6 ENST00000671668.1 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr16_-_75267988 | 0.19 |
ENST00000393422.6
|
BCAR1
|
BCAR1 scaffold protein, Cas family member |
| chrX_+_123960859 | 0.19 |
ENST00000458700.5
|
STAG2
|
stromal antigen 2 |
| chr6_+_125203639 | 0.18 |
ENST00000392482.6
|
TPD52L1
|
TPD52 like 1 |
| chr22_+_38057200 | 0.18 |
ENST00000404072.7
ENST00000424694.5 |
PICK1
|
protein interacting with PRKCA 1 |
| chr9_+_93289187 | 0.18 |
ENST00000453718.2
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr4_+_75556048 | 0.18 |
ENST00000616557.1
ENST00000435974.2 ENST00000311623.9 |
ODAPH
|
odontogenesis associated phosphoprotein |
| chr6_+_147508645 | 0.17 |
ENST00000367474.2
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr15_-_28174423 | 0.17 |
ENST00000569772.1
|
HERC2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr9_-_14322320 | 0.17 |
ENST00000606230.2
|
NFIB
|
nuclear factor I B |
| chr5_+_35852695 | 0.17 |
ENST00000508941.5
|
IL7R
|
interleukin 7 receptor |
| chr6_-_31958935 | 0.16 |
ENST00000441998.5
ENST00000444811.6 ENST00000375429.8 |
NELFE
|
negative elongation factor complex member E |
| chr3_-_56468346 | 0.16 |
ENST00000288221.11
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr15_-_34754989 | 0.16 |
ENST00000290374.5
|
GJD2
|
gap junction protein delta 2 |
| chr7_-_5959083 | 0.16 |
ENST00000539903.5
|
RSPH10B
|
radial spoke head 10 homolog B |
| chr10_-_14554141 | 0.16 |
ENST00000496330.5
|
FAM107B
|
family with sequence similarity 107 member B |
| chr2_+_135531460 | 0.16 |
ENST00000683871.1
ENST00000409478.5 ENST00000264160.8 ENST00000438014.5 |
R3HDM1
|
R3H domain containing 1 |
| chr8_-_140800535 | 0.16 |
ENST00000521986.5
ENST00000523539.5 |
PTK2
|
protein tyrosine kinase 2 |
| chr10_+_122192442 | 0.16 |
ENST00000514539.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr2_-_206159509 | 0.16 |
ENST00000423725.5
|
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr11_-_114595750 | 0.15 |
ENST00000424261.6
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4 |
| chr12_+_25052220 | 0.15 |
ENST00000550945.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr12_+_25052634 | 0.15 |
ENST00000548766.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chrX_+_41689006 | 0.15 |
ENST00000378138.5
ENST00000620846.1 ENST00000649219.1 |
GPR34
|
G protein-coupled receptor 34 |
| chr5_-_108382080 | 0.15 |
ENST00000542267.7
|
FBXL17
|
F-box and leucine rich repeat protein 17 |
| chr1_-_66801276 | 0.15 |
ENST00000304526.3
|
INSL5
|
insulin like 5 |
| chr12_-_262828 | 0.15 |
ENST00000343164.9
ENST00000436453.1 ENST00000445055.6 ENST00000546319.5 |
SLC6A13
|
solute carrier family 6 member 13 |
| chr13_-_46390042 | 0.15 |
ENST00000389908.7
|
RUBCNL
|
rubicon like autophagy enhancer |
| chr12_-_6493718 | 0.15 |
ENST00000543959.5
ENST00000543164.5 ENST00000543703.1 ENST00000537701.5 |
MRPL51
|
mitochondrial ribosomal protein L51 |
| chr2_-_206159410 | 0.15 |
ENST00000457011.5
ENST00000440274.5 ENST00000432169.5 ENST00000233190.11 |
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr20_+_43514492 | 0.14 |
ENST00000373135.8
ENST00000373134.5 |
L3MBTL1
|
L3MBTL histone methyl-lysine binding protein 1 |
| chr12_-_84892120 | 0.14 |
ENST00000680379.1
|
SLC6A15
|
solute carrier family 6 member 15 |
| chr17_-_81683659 | 0.14 |
ENST00000574938.5
ENST00000570561.5 ENST00000573392.5 ENST00000576135.5 ENST00000573715.2 |
ARL16
|
ADP ribosylation factor like GTPase 16 |
| chr22_+_31122923 | 0.14 |
ENST00000620191.4
ENST00000412277.6 ENST00000412985.5 ENST00000331075.10 ENST00000420017.5 ENST00000400294.6 ENST00000405300.5 ENST00000404390.7 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr17_+_64507029 | 0.14 |
ENST00000553412.5
|
CEP95
|
centrosomal protein 95 |
| chr14_-_24263162 | 0.13 |
ENST00000206765.11
ENST00000544573.5 |
TGM1
|
transglutaminase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.5 | 2.0 | GO:0002086 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.4 | 3.5 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.3 | 24.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 0.7 | GO:1902598 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.2 | 0.5 | GO:0002543 | activation of blood coagulation via clotting cascade(GO:0002543) |
| 0.2 | 1.6 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.2 | 0.6 | GO:1901383 | negative regulation of chorionic trophoblast cell proliferation(GO:1901383) |
| 0.2 | 0.9 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 5.5 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 1.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.2 | 0.5 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 0.6 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.8 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 1.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 1.4 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.7 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.4 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 9.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.3 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.3 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.1 | 0.2 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.1 | 0.4 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.5 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.7 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.1 | 0.2 | GO:0097115 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.1 | 1.0 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 0.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.3 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.5 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.5 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:2000374 | regulation of oxygen metabolic process(GO:2000374) |
| 0.0 | 0.8 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.9 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.5 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 1.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.9 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.9 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.6 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.5 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.3 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.3 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.4 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.8 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 1.2 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.3 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 0.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.3 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 1.5 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.2 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 24.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 2.0 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.3 | 1.0 | GO:1905103 | integral component of lysosomal membrane(GO:1905103) |
| 0.1 | 0.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 1.0 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.1 | 0.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.6 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.1 | 0.4 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 0.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.5 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 1.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.8 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 3.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.9 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.1 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.6 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 1.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.7 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.6 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 4.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 5.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.5 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 2.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.3 | 24.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.3 | 5.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 0.7 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.2 | 1.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.6 | GO:0038100 | nodal binding(GO:0038100) |
| 0.2 | 1.7 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.4 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.8 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 1.6 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.4 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.4 | GO:0030379 | neurotensin receptor activity, non-G-protein coupled(GO:0030379) |
| 0.1 | 0.7 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 0.5 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 1.0 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 1.4 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 0.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 1.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 12.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.3 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.5 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.6 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.9 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 4.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.5 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.9 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 1.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.9 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |