Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for NKX3-2
Z-value: 0.94
Transcription factors associated with NKX3-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX3-2
|
ENSG00000109705.8 | NK3 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX3-2 | hg38_v1_chr4_-_13544506_13544513 | -0.10 | 5.8e-01 | Click! |
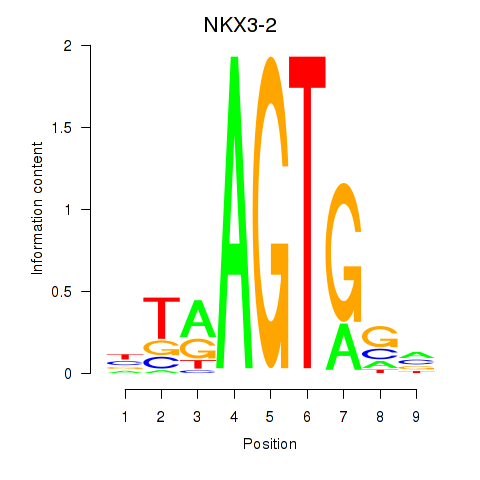
Activity profile of NKX3-2 motif
Sorted Z-values of NKX3-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_22161788 | 2.91 |
ENST00000521315.5
ENST00000437090.6 ENST00000679463.1 ENST00000520605.5 ENST00000522109.5 ENST00000524255.5 ENST00000523296.1 |
SFTPC
|
surfactant protein C |
| chr8_+_22161655 | 2.64 |
ENST00000318561.7
|
SFTPC
|
surfactant protein C |
| chr14_-_106557465 | 2.14 |
ENST00000390625.3
|
IGHV3-49
|
immunoglobulin heavy variable 3-49 |
| chr7_-_120858066 | 1.94 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12 |
| chr7_-_99976017 | 1.94 |
ENST00000411734.1
ENST00000292401.9 |
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr6_-_32763522 | 1.94 |
ENST00000435145.6
ENST00000437316.7 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr14_-_106639589 | 1.90 |
ENST00000390630.3
|
IGHV4-61
|
immunoglobulin heavy variable 4-61 |
| chr3_+_194136138 | 1.78 |
ENST00000232424.4
|
HES1
|
hes family bHLH transcription factor 1 |
| chr14_-_106579223 | 1.78 |
ENST00000390626.2
|
IGHV5-51
|
immunoglobulin heavy variable 5-51 |
| chr5_+_147878703 | 1.78 |
ENST00000296694.5
|
SCGB3A2
|
secretoglobin family 3A member 2 |
| chr6_-_30932147 | 1.77 |
ENST00000359086.4
|
SFTA2
|
surfactant associated 2 |
| chr16_-_1230089 | 1.71 |
ENST00000612142.1
ENST00000606293.5 |
TPSB2
|
tryptase beta 2 |
| chr7_-_99971845 | 1.71 |
ENST00000419575.1
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr2_+_102311546 | 1.61 |
ENST00000233954.6
ENST00000447231.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr6_-_32763466 | 1.60 |
ENST00000427449.1
ENST00000411527.5 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr11_-_102530738 | 1.55 |
ENST00000260227.5
|
MMP7
|
matrix metallopeptidase 7 |
| chr19_+_16888991 | 1.55 |
ENST00000248076.4
|
F2RL3
|
F2R like thrombin or trypsin receptor 3 |
| chr14_-_106507476 | 1.53 |
ENST00000390621.3
|
IGHV1-45
|
immunoglobulin heavy variable 1-45 |
| chr2_+_102311502 | 1.48 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr5_+_69415065 | 1.45 |
ENST00000647531.1
ENST00000645446.1 ENST00000325631.10 ENST00000454295.6 |
MARVELD2
|
MARVEL domain containing 2 |
| chr10_+_5048748 | 1.44 |
ENST00000602997.5
ENST00000439082.7 |
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr11_-_120138104 | 1.42 |
ENST00000341846.10
|
TRIM29
|
tripartite motif containing 29 |
| chr5_+_79111802 | 1.39 |
ENST00000524080.1
ENST00000274353.10 |
BHMT
|
betaine--homocysteine S-methyltransferase |
| chr4_+_42397473 | 1.38 |
ENST00000319234.5
|
SHISA3
|
shisa family member 3 |
| chr14_-_106374129 | 1.30 |
ENST00000390616.2
|
IGHV4-34
|
immunoglobulin heavy variable 4-34 |
| chr17_+_4498866 | 1.30 |
ENST00000329078.8
|
SPNS2
|
sphingolipid transporter 2 |
| chr2_-_88861563 | 1.29 |
ENST00000624935.3
ENST00000390241.3 |
ENSG00000240040.6
IGKJ2
|
novel transcript immunoglobulin kappa joining 2 |
| chr14_-_106627685 | 1.28 |
ENST00000390629.3
|
IGHV4-59
|
immunoglobulin heavy variable 4-59 |
| chr13_-_110242694 | 1.24 |
ENST00000648989.1
ENST00000647797.1 ENST00000648966.1 ENST00000649484.1 ENST00000648695.1 ENST00000650115.1 ENST00000650566.1 |
COL4A1
|
collagen type IV alpha 1 chain |
| chr3_-_51974001 | 1.24 |
ENST00000489595.6
ENST00000461108.5 ENST00000395008.6 ENST00000361143.10 ENST00000525795.1 ENST00000488257.2 |
PCBP4
ABHD14B
ENSG00000272762.5
|
poly(rC) binding protein 4 abhydrolase domain containing 14B novel transcript |
| chr10_-_96271508 | 1.22 |
ENST00000427367.6
ENST00000413476.6 ENST00000371176.6 |
BLNK
|
B cell linker |
| chr12_-_50222348 | 1.15 |
ENST00000552823.5
ENST00000552909.5 |
LIMA1
|
LIM domain and actin binding 1 |
| chr1_-_153544997 | 1.15 |
ENST00000368715.5
|
S100A4
|
S100 calcium binding protein A4 |
| chr3_+_132597260 | 1.13 |
ENST00000249887.3
|
ACKR4
|
atypical chemokine receptor 4 |
| chr10_-_5977492 | 1.13 |
ENST00000530685.5
ENST00000397255.7 ENST00000379971.5 ENST00000528354.5 ENST00000397250.6 ENST00000429135.2 |
IL15RA
|
interleukin 15 receptor subunit alpha |
| chr10_+_5094405 | 1.11 |
ENST00000380554.5
|
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr11_+_60455839 | 1.09 |
ENST00000532491.5
ENST00000532073.5 ENST00000345732.9 ENST00000534668.6 ENST00000528313.1 ENST00000533306.6 ENST00000674194.1 |
MS4A1
|
membrane spanning 4-domains A1 |
| chr6_-_32589833 | 1.09 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr7_-_131556602 | 1.08 |
ENST00000322985.9
ENST00000378555.8 |
PODXL
|
podocalyxin like |
| chr12_-_21774688 | 1.06 |
ENST00000240662.3
|
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8 |
| chr6_-_11382247 | 1.05 |
ENST00000397378.7
ENST00000513989.5 ENST00000508546.5 ENST00000504387.5 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr5_+_40909490 | 1.05 |
ENST00000313164.10
|
C7
|
complement C7 |
| chr12_-_21775045 | 1.04 |
ENST00000667884.1
|
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8 |
| chr16_+_57372481 | 1.02 |
ENST00000006053.7
|
CX3CL1
|
C-X3-C motif chemokine ligand 1 |
| chr14_+_22547495 | 1.00 |
ENST00000611116.2
|
TRAC
|
T cell receptor alpha constant |
| chr1_-_149842736 | 0.98 |
ENST00000369159.2
|
H2AC18
|
H2A clustered histone 18 |
| chr11_-_26572102 | 0.98 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated |
| chr14_-_105940235 | 0.98 |
ENST00000390593.2
|
IGHV6-1
|
immunoglobulin heavy variable 6-1 |
| chr16_+_57372465 | 0.98 |
ENST00000563383.1
|
CX3CL1
|
C-X3-C motif chemokine ligand 1 |
| chr8_-_94262308 | 0.97 |
ENST00000297596.3
ENST00000396194.6 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr4_+_70637662 | 0.96 |
ENST00000472597.1
ENST00000472903.5 |
ENAM
ENSG00000286848.1
|
enamelin novel transcript |
| chr3_+_186996444 | 0.96 |
ENST00000676633.1
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr14_-_106005574 | 0.96 |
ENST00000390595.3
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr14_+_61697622 | 0.96 |
ENST00000539097.2
|
HIF1A
|
hypoxia inducible factor 1 subunit alpha |
| chrX_-_108438407 | 0.96 |
ENST00000334504.12
ENST00000621266.4 ENST00000538570.5 ENST00000394872.6 ENST00000545689.2 |
COL4A6
|
collagen type IV alpha 6 chain |
| chr11_-_5243644 | 0.96 |
ENST00000643122.1
|
HBD
|
hemoglobin subunit delta |
| chr4_+_155903688 | 0.95 |
ENST00000536354.3
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr1_-_204213943 | 0.95 |
ENST00000308302.4
|
GOLT1A
|
golgi transport 1A |
| chr7_+_116210501 | 0.95 |
ENST00000455989.1
ENST00000358204.9 |
TES
|
testin LIM domain protein |
| chr10_-_5966327 | 0.94 |
ENST00000532039.5
|
IL15RA
|
interleukin 15 receptor subunit alpha |
| chrX_+_106693751 | 0.94 |
ENST00000418562.5
|
RNF128
|
ring finger protein 128 |
| chr15_-_43220989 | 0.94 |
ENST00000540029.5
ENST00000441366.7 ENST00000648595.1 |
EPB42
|
erythrocyte membrane protein band 4.2 |
| chr19_+_40991274 | 0.93 |
ENST00000324071.10
|
CYP2B6
|
cytochrome P450 family 2 subfamily B member 6 |
| chr11_-_59866478 | 0.93 |
ENST00000257264.4
|
TCN1
|
transcobalamin 1 |
| chr14_-_106012390 | 0.91 |
ENST00000455737.2
|
IGHV4-4
|
immunoglobulin heavy variable 4-4 |
| chr11_-_76669985 | 0.90 |
ENST00000407242.6
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr1_-_36482904 | 0.89 |
ENST00000373106.6
|
CSF3R
|
colony stimulating factor 3 receptor |
| chr1_-_159925496 | 0.89 |
ENST00000368097.9
|
TAGLN2
|
transgelin 2 |
| chr7_+_116222804 | 0.88 |
ENST00000393481.6
|
TES
|
testin LIM domain protein |
| chr9_-_92482499 | 0.88 |
ENST00000375544.7
|
ASPN
|
asporin |
| chr12_+_49961864 | 0.88 |
ENST00000293599.7
|
AQP5
|
aquaporin 5 |
| chr17_+_18952158 | 0.87 |
ENST00000395647.6
ENST00000417251.6 ENST00000395645.4 |
SLC5A10
|
solute carrier family 5 member 10 |
| chr11_-_26572254 | 0.87 |
ENST00000529533.6
|
MUC15
|
mucin 15, cell surface associated |
| chr12_+_80099535 | 0.85 |
ENST00000646859.1
ENST00000547103.7 |
OTOGL
|
otogelin like |
| chr12_-_14929116 | 0.85 |
ENST00000540097.1
|
ERP27
|
endoplasmic reticulum protein 27 |
| chr9_-_129178247 | 0.84 |
ENST00000372491.4
|
IER5L
|
immediate early response 5 like |
| chr4_+_168711416 | 0.83 |
ENST00000649826.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_+_81800906 | 0.83 |
ENST00000674393.1
ENST00000674208.1 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr14_-_106324743 | 0.83 |
ENST00000390612.3
|
IGHV4-28
|
immunoglobulin heavy variable 4-28 |
| chr15_-_90233902 | 0.81 |
ENST00000328649.11
ENST00000650306.1 |
CIB1
|
calcium and integrin binding 1 |
| chr11_-_74697694 | 0.81 |
ENST00000529912.5
|
CHRDL2
|
chordin like 2 |
| chrX_+_106693838 | 0.81 |
ENST00000324342.7
|
RNF128
|
ring finger protein 128 |
| chr19_-_46784905 | 0.81 |
ENST00000594991.5
|
SLC1A5
|
solute carrier family 1 member 5 |
| chr11_-_128867364 | 0.79 |
ENST00000440599.6
ENST00000324036.7 |
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr11_-_128867268 | 0.79 |
ENST00000392665.6
ENST00000392666.6 |
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr19_+_41877267 | 0.78 |
ENST00000221972.8
ENST00000597454.1 ENST00000444740.2 |
CD79A
|
CD79a molecule |
| chr10_+_8053604 | 0.78 |
ENST00000481743.2
|
GATA3
|
GATA binding protein 3 |
| chr12_-_121858849 | 0.77 |
ENST00000289004.8
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase |
| chr12_-_21775581 | 0.77 |
ENST00000537950.1
ENST00000665145.1 |
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8 |
| chr17_-_48430205 | 0.76 |
ENST00000336915.11
ENST00000584924.5 |
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr9_-_22009272 | 0.76 |
ENST00000380142.5
ENST00000276925.7 |
CDKN2B
|
cyclin dependent kinase inhibitor 2B |
| chr2_+_79120474 | 0.76 |
ENST00000233735.2
|
REG1A
|
regenerating family member 1 alpha |
| chr1_+_149851053 | 0.75 |
ENST00000607355.2
|
H2AC19
|
H2A clustered histone 19 |
| chr9_+_113349514 | 0.75 |
ENST00000374183.5
|
BSPRY
|
B-box and SPRY domain containing |
| chr14_+_20688756 | 0.75 |
ENST00000397990.5
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin ribonuclease A family member 4 |
| chr10_-_7666955 | 0.74 |
ENST00000397146.7
ENST00000397145.6 |
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain 5 |
| chr17_+_70075215 | 0.73 |
ENST00000283936.5
ENST00000615244.4 ENST00000392671.6 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr9_-_127847117 | 0.72 |
ENST00000480266.5
|
ENG
|
endoglin |
| chr4_+_147480917 | 0.72 |
ENST00000324300.10
ENST00000358556.8 ENST00000511804.5 ENST00000648866.1 |
EDNRA
|
endothelin receptor type A |
| chr16_+_57372525 | 0.71 |
ENST00000564948.1
|
CX3CL1
|
C-X3-C motif chemokine ligand 1 |
| chr9_-_92482350 | 0.71 |
ENST00000375543.2
|
ASPN
|
asporin |
| chr10_-_47484081 | 0.71 |
ENST00000583448.2
ENST00000583874.5 ENST00000585281.6 |
ANXA8
|
annexin A8 |
| chr16_-_88785210 | 0.71 |
ENST00000301015.14
|
PIEZO1
|
piezo type mechanosensitive ion channel component 1 |
| chr13_+_110424798 | 0.69 |
ENST00000619688.2
|
COL4A2
|
collagen type IV alpha 2 chain |
| chr15_+_81182579 | 0.69 |
ENST00000302987.9
|
IL16
|
interleukin 16 |
| chr8_-_48921419 | 0.68 |
ENST00000020945.4
|
SNAI2
|
snail family transcriptional repressor 2 |
| chr5_-_176899332 | 0.68 |
ENST00000292432.10
|
HK3
|
hexokinase 3 |
| chr6_-_31729785 | 0.67 |
ENST00000416410.6
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr20_+_43916142 | 0.67 |
ENST00000423191.6
ENST00000372999.5 |
TOX2
|
TOX high mobility group box family member 2 |
| chr6_-_31729478 | 0.67 |
ENST00000436437.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr6_-_138572523 | 0.67 |
ENST00000427025.6
|
NHSL1
|
NHS like 1 |
| chr11_+_34621109 | 0.67 |
ENST00000450654.6
|
EHF
|
ETS homologous factor |
| chr6_-_47042306 | 0.66 |
ENST00000371253.7
|
ADGRF1
|
adhesion G protein-coupled receptor F1 |
| chr7_+_80638633 | 0.66 |
ENST00000447544.7
ENST00000482059.6 |
CD36
|
CD36 molecule |
| chr1_-_24930263 | 0.65 |
ENST00000308873.11
|
RUNX3
|
RUNX family transcription factor 3 |
| chr12_-_50222694 | 0.65 |
ENST00000552783.5
|
LIMA1
|
LIM domain and actin binding 1 |
| chr4_+_88007624 | 0.65 |
ENST00000237596.7
|
PKD2
|
polycystin 2, transient receptor potential cation channel |
| chr11_+_35201083 | 0.65 |
ENST00000526553.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr3_-_149670877 | 0.64 |
ENST00000475579.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr11_-_120123026 | 0.64 |
ENST00000533302.5
|
TRIM29
|
tripartite motif containing 29 |
| chr1_+_86424154 | 0.64 |
ENST00000370565.5
|
CLCA2
|
chloride channel accessory 2 |
| chr14_-_106622837 | 0.63 |
ENST00000390628.3
|
IGHV1-58
|
immunoglobulin heavy variable 1-58 |
| chr2_+_176151543 | 0.62 |
ENST00000306324.4
|
HOXD4
|
homeobox D4 |
| chr5_-_150412743 | 0.62 |
ENST00000353334.11
ENST00000009530.12 ENST00000377795.7 |
CD74
|
CD74 molecule |
| chr16_+_83953232 | 0.62 |
ENST00000565123.5
ENST00000393306.6 |
OSGIN1
|
oxidative stress induced growth inhibitor 1 |
| chr9_+_109780312 | 0.62 |
ENST00000483909.5
ENST00000413420.5 ENST00000302798.7 |
PALM2AKAP2
|
PALM2 and AKAP2 fusion |
| chr3_-_100832300 | 0.62 |
ENST00000478235.5
ENST00000471901.5 |
ABI3BP
|
ABI family member 3 binding protein |
| chr18_+_44680093 | 0.62 |
ENST00000426838.8
ENST00000677068.1 |
SETBP1
|
SET binding protein 1 |
| chr6_-_31730198 | 0.62 |
ENST00000375787.6
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr7_+_128937917 | 0.62 |
ENST00000357234.10
ENST00000613821.4 ENST00000477535.5 ENST00000479582.5 ENST00000464557.5 ENST00000402030.6 |
IRF5
|
interferon regulatory factor 5 |
| chr7_+_150450623 | 0.61 |
ENST00000307271.4
|
GIMAP8
|
GTPase, IMAP family member 8 |
| chr11_-_107018462 | 0.61 |
ENST00000526355.7
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr12_+_6385119 | 0.61 |
ENST00000541102.1
|
LTBR
|
lymphotoxin beta receptor |
| chr3_-_100840109 | 0.60 |
ENST00000533795.5
|
ABI3BP
|
ABI family member 3 binding protein |
| chr11_+_34621065 | 0.60 |
ENST00000257831.8
|
EHF
|
ETS homologous factor |
| chr14_-_106108453 | 0.60 |
ENST00000632950.2
|
IGHV5-10-1
|
immunoglobulin heavy variable 5-10-1 |
| chr19_+_852295 | 0.58 |
ENST00000263621.2
|
ELANE
|
elastase, neutrophil expressed |
| chr1_-_182604379 | 0.58 |
ENST00000367558.6
|
RGS16
|
regulator of G protein signaling 16 |
| chr2_+_85584402 | 0.58 |
ENST00000306384.5
|
VAMP5
|
vesicle associated membrane protein 5 |
| chr7_+_76510528 | 0.58 |
ENST00000334348.8
|
UPK3B
|
uroplakin 3B |
| chr4_+_147481085 | 0.57 |
ENST00000651419.1
|
EDNRA
|
endothelin receptor type A |
| chr9_-_92482461 | 0.57 |
ENST00000651738.1
|
ASPN
|
asporin |
| chr4_+_108650644 | 0.56 |
ENST00000512478.2
|
OSTC
|
oligosaccharyltransferase complex non-catalytic subunit |
| chr5_+_56815534 | 0.55 |
ENST00000399503.4
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1 |
| chr1_+_162381703 | 0.54 |
ENST00000458626.4
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr7_+_80638662 | 0.54 |
ENST00000394788.7
|
CD36
|
CD36 molecule |
| chr17_+_7579491 | 0.53 |
ENST00000380498.10
ENST00000584502.1 ENST00000250092.11 |
CD68
|
CD68 molecule |
| chr2_-_27071628 | 0.53 |
ENST00000447619.5
ENST00000429985.1 ENST00000456793.2 |
OST4
|
oligosaccharyltransferase complex subunit 4, non-catalytic |
| chr2_-_88860913 | 0.52 |
ENST00000390239.2
|
IGKJ4
|
immunoglobulin kappa joining 4 |
| chr14_-_106875069 | 0.52 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr15_-_90234046 | 0.52 |
ENST00000612800.1
|
CIB1
|
calcium and integrin binding 1 |
| chr5_+_76609091 | 0.51 |
ENST00000514001.5
ENST00000396234.7 ENST00000509074.5 ENST00000502745.5 |
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr17_-_76141240 | 0.51 |
ENST00000322957.7
|
FOXJ1
|
forkhead box J1 |
| chr11_-_26572130 | 0.50 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr15_-_53733103 | 0.50 |
ENST00000559418.5
|
WDR72
|
WD repeat domain 72 |
| chr17_+_70075317 | 0.50 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr1_-_173917281 | 0.50 |
ENST00000367698.4
|
SERPINC1
|
serpin family C member 1 |
| chr18_+_44680875 | 0.49 |
ENST00000649279.2
ENST00000677699.1 |
SETBP1
|
SET binding protein 1 |
| chr3_-_42875871 | 0.49 |
ENST00000316161.6
ENST00000437102.1 |
CYP8B1
|
cytochrome P450 family 8 subfamily B member 1 |
| chr2_+_237486391 | 0.49 |
ENST00000429898.5
ENST00000410032.5 |
MLPH
|
melanophilin |
| chr14_+_20999255 | 0.49 |
ENST00000554422.5
ENST00000298681.5 |
SLC39A2
|
solute carrier family 39 member 2 |
| chr4_-_118352967 | 0.49 |
ENST00000296498.3
|
PRSS12
|
serine protease 12 |
| chr8_-_48921735 | 0.49 |
ENST00000396822.6
|
SNAI2
|
snail family transcriptional repressor 2 |
| chr12_-_57767057 | 0.48 |
ENST00000228606.9
|
CYP27B1
|
cytochrome P450 family 27 subfamily B member 1 |
| chr11_-_120138031 | 0.47 |
ENST00000627238.1
|
TRIM29
|
tripartite motif containing 29 |
| chr6_-_32853813 | 0.47 |
ENST00000643049.2
|
TAP1
|
transporter 1, ATP binding cassette subfamily B member |
| chr19_-_49325181 | 0.47 |
ENST00000454748.7
ENST00000335875.9 ENST00000598828.1 |
SLC6A16
|
solute carrier family 6 member 16 |
| chr18_+_23884773 | 0.46 |
ENST00000586751.5
|
LAMA3
|
laminin subunit alpha 3 |
| chr1_-_8015633 | 0.45 |
ENST00000467067.1
|
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr5_+_69415382 | 0.45 |
ENST00000512803.5
|
MARVELD2
|
MARVEL domain containing 2 |
| chr17_-_81656532 | 0.45 |
ENST00000331056.10
|
PDE6G
|
phosphodiesterase 6G |
| chr1_+_23959109 | 0.45 |
ENST00000471915.5
|
PNRC2
|
proline rich nuclear receptor coactivator 2 |
| chr5_-_1345084 | 0.45 |
ENST00000320895.10
|
CLPTM1L
|
CLPTM1 like |
| chr19_-_46023046 | 0.44 |
ENST00000008938.5
|
PGLYRP1
|
peptidoglycan recognition protein 1 |
| chr11_+_13962676 | 0.43 |
ENST00000576479.4
|
SPON1
|
spondin 1 |
| chr2_-_38751350 | 0.43 |
ENST00000409276.5
ENST00000313117.11 ENST00000446327.6 |
SRSF7
|
serine and arginine rich splicing factor 7 |
| chr1_-_204494752 | 0.43 |
ENST00000684373.1
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr11_+_93741591 | 0.43 |
ENST00000528288.5
ENST00000617482.4 ENST00000540113.5 |
C11orf54
|
chromosome 11 open reading frame 54 |
| chr12_+_49741544 | 0.43 |
ENST00000549966.5
ENST00000547832.5 ENST00000547187.5 ENST00000548894.5 ENST00000546914.5 ENST00000552699.5 ENST00000267115.10 |
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr3_-_146544578 | 0.43 |
ENST00000342435.9
ENST00000448787.6 |
PLSCR1
|
phospholipid scramblase 1 |
| chr4_+_48016764 | 0.42 |
ENST00000295461.10
|
NIPAL1
|
NIPA like domain containing 1 |
| chr22_-_37244417 | 0.42 |
ENST00000405484.5
ENST00000441619.5 ENST00000406508.5 |
RAC2
|
Rac family small GTPase 2 |
| chr2_-_70553638 | 0.42 |
ENST00000444975.5
ENST00000445399.5 ENST00000295400.11 ENST00000418333.6 |
TGFA
|
transforming growth factor alpha |
| chr6_-_116060859 | 0.41 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr17_-_45410414 | 0.41 |
ENST00000532038.5
ENST00000528677.1 |
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr11_+_63938971 | 0.41 |
ENST00000539656.5
ENST00000377793.9 |
NAA40
|
N-alpha-acetyltransferase 40, NatD catalytic subunit |
| chr10_+_100998676 | 0.41 |
ENST00000481129.1
|
LZTS2
|
leucine zipper tumor suppressor 2 |
| chr4_+_17577487 | 0.41 |
ENST00000606142.5
|
LAP3
|
leucine aminopeptidase 3 |
| chr9_+_35161951 | 0.40 |
ENST00000617908.4
ENST00000619578.4 |
UNC13B
|
unc-13 homolog B |
| chr5_+_111071710 | 0.40 |
ENST00000344895.4
|
TSLP
|
thymic stromal lymphopoietin |
| chr7_+_142615710 | 0.40 |
ENST00000611520.1
|
TRBV18
|
T cell receptor beta variable 18 |
| chr1_+_93079264 | 0.40 |
ENST00000370298.9
ENST00000370303.4 |
MTF2
|
metal response element binding transcription factor 2 |
| chr3_+_122184233 | 0.40 |
ENST00000638421.1
ENST00000498619.4 |
CASR
|
calcium sensing receptor |
| chr9_+_35162000 | 0.39 |
ENST00000396787.5
ENST00000378495.7 ENST00000635942.1 ENST00000378496.8 |
UNC13B
|
unc-13 homolog B |
| chr3_+_189631373 | 0.39 |
ENST00000264731.8
ENST00000418709.6 ENST00000320472.9 ENST00000392460.7 ENST00000440651.6 |
TP63
|
tumor protein p63 |
| chr2_+_157257687 | 0.39 |
ENST00000259056.5
|
GALNT5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr20_-_2840623 | 0.39 |
ENST00000360652.7
ENST00000448755.5 |
PCED1A
|
PC-esterase domain containing 1A |
| chr21_-_34887148 | 0.39 |
ENST00000399240.5
|
RUNX1
|
RUNX family transcription factor 1 |
| chr19_+_40807112 | 0.38 |
ENST00000595621.1
ENST00000595051.1 |
EGLN2
|
egl-9 family hypoxia inducible factor 2 |
| chr3_-_146544701 | 0.38 |
ENST00000487389.5
|
PLSCR1
|
phospholipid scramblase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX3-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0051041 | positive regulation of calcium-independent cell-cell adhesion(GO:0051041) |
| 0.9 | 2.6 | GO:0016107 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.6 | 1.8 | GO:0045608 | auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.5 | 1.4 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.4 | 1.8 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.4 | 1.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.4 | 3.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 1.3 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.3 | 1.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 1.3 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.3 | 1.0 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.3 | 1.0 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.3 | 1.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.3 | 0.8 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.3 | 2.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.3 | 0.8 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.2 | 1.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.2 | 0.7 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.2 | 0.7 | GO:0071464 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) cellular response to hydrostatic pressure(GO:0071464) |
| 0.2 | 0.6 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.2 | 0.8 | GO:1903803 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.2 | 0.8 | GO:2000701 | regulation of cell proliferation involved in mesonephros development(GO:2000606) negative regulation of cell proliferation involved in mesonephros development(GO:2000607) fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000699) glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000701) regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000702) negative regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000703) regulation of glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000733) negative regulation of glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000734) |
| 0.2 | 1.3 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.2 | 0.7 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.2 | 0.9 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 0.5 | GO:0070564 | positive regulation of vitamin D receptor signaling pathway(GO:0070564) |
| 0.2 | 1.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.2 | 13.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.4 | GO:0032824 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 1.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 1.7 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 1.9 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 0.5 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.7 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.3 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.7 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.1 | 0.4 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.6 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.1 | 0.3 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.1 | 0.3 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 2.0 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.1 | 0.3 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.1 | 2.9 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.1 | 1.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.5 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 0.2 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.1 | 3.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.3 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.7 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 | 1.0 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.2 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.3 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.1 | 0.2 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.1 | 0.4 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.1 | 1.5 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 1.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.0 | 1.5 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.6 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 1.2 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.2 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.4 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.3 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 1.9 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 1.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.7 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 4.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0051511 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.1 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 1.0 | GO:0015671 | gas transport(GO:0015669) oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.6 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.6 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.2 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 1.2 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.2 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) |
| 0.0 | 0.2 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.1 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.6 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.4 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 2.8 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 4.0 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.2 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 2.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.4 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.2 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.3 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 1.8 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.5 | GO:0002438 | acute inflammatory response to antigenic stimulus(GO:0002438) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.5 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.4 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 2.1 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.6 | GO:0070232 | regulation of T cell apoptotic process(GO:0070232) |
| 0.0 | 0.6 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 1.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.4 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.7 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:1901254 | modulation by host of viral RNA genome replication(GO:0044830) negative regulation of long term synaptic depression(GO:1900453) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.0 | 0.1 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.0 | 0.5 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.6 | GO:0032735 | positive regulation of interleukin-12 production(GO:0032735) |
| 0.0 | 0.5 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 2.0 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.4 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.2 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.6 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.2 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.1 | GO:0019682 | fructose catabolic process(GO:0006001) glyceraldehyde-3-phosphate metabolic process(GO:0019682) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.7 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.3 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.4 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.3 | 5.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 2.9 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 0.7 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.2 | 0.7 | GO:0016590 | ACF complex(GO:0016590) |
| 0.2 | 12.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 2.9 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 2.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 0.8 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.1 | 0.7 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.1 | 1.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.8 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.7 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.8 | GO:0044305 | early phagosome(GO:0032009) calyx of Held(GO:0044305) |
| 0.1 | 0.4 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 2.0 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 1.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 1.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.2 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 1.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.6 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 1.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 1.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.1 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 1.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 3.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 2.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 4.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.9 | 2.7 | GO:0031737 | CX3C chemokine receptor binding(GO:0031737) |
| 0.9 | 2.6 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.5 | 2.9 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.3 | 1.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.2 | 2.0 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 3.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 1.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 1.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 1.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.2 | 1.3 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 4.0 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.2 | 12.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 0.8 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.2 | 1.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 1.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.7 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.7 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.4 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 1.0 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 0.4 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 0.8 | GO:0023029 | MHC class Ib protein binding(GO:0023029) TAP2 binding(GO:0046979) |
| 0.1 | 1.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 1.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.0 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.7 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 0.7 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.8 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.8 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.3 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.4 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 2.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 1.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.3 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.3 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.6 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.6 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 2.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.9 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 1.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.4 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.6 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 1.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 1.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 1.1 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.2 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 4.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 3.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 3.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.8 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.3 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 3.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0018479 | androgen binding(GO:0005497) benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0019871 | potassium channel inhibitor activity(GO:0019870) sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 1.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 3.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 9.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 6.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 4.8 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 1.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.9 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 0.9 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 0.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 2.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 2.8 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 1.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.9 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.7 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.1 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |